Search Count: 666
 |
Nanoparticle Crystal Structure Of A Thermostabilized Mutant Rv1498A Flavoprotein From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN |
 |
Nanoparticle Crystal Structure Of A Thermostabilized Mutant Of An As-Isolated Ftna (Ferritin) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: METAL BINDING PROTEIN |
 |
Nanoparticle Crystal Structure Of A Thermostabilized Mutant E.Coli Ferritin Ecftna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
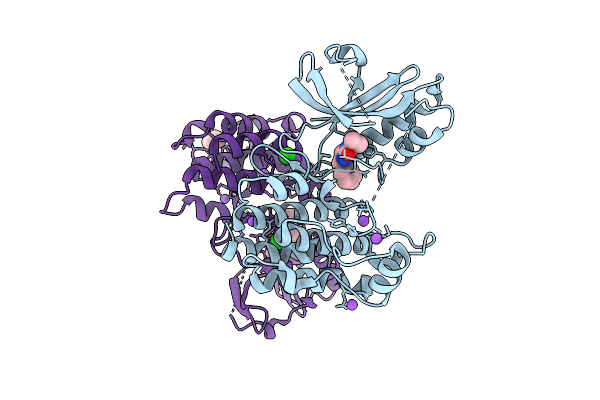 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE/Inhibitor Ligands: A1CNU, BR, CL, NA, TME |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: Transferase/Inhibitor Ligands: A1CNV |
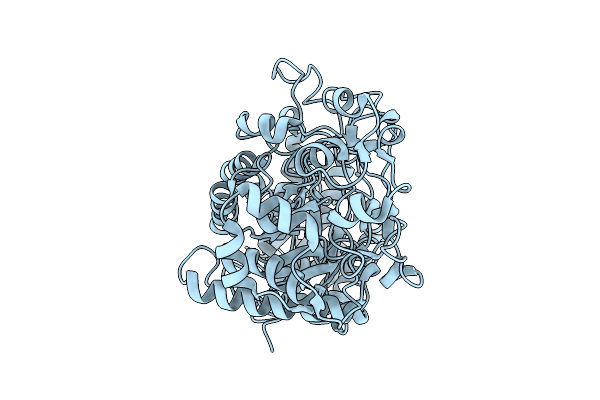 |
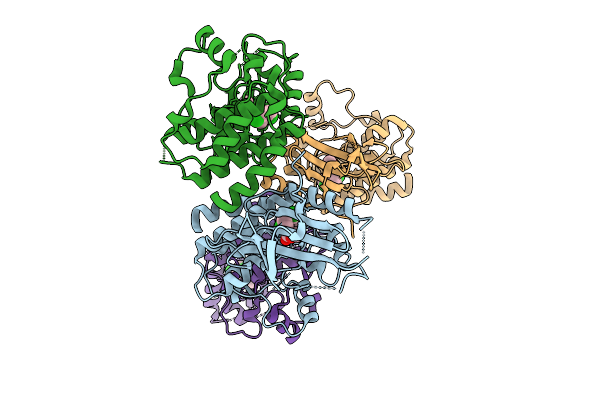
Organism: Ruminococcus bromii l2-63
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
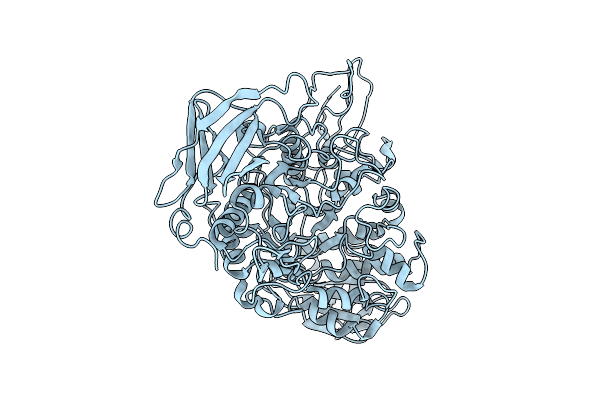 |
Organism: Ruminococcus bromii l2-63
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
 |
Organism: Ruminococcus bromii l2-63
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
 |
Organism: Ruminococcus bromii l2-63
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE |
 |
Organism: Glutamicibacter protophormiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE Ligands: CA, DUC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |

