Search Count: 2,924
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structures And Mechanism Of The Nicotinamide Nucleotide Transhydrogenase From E. Coli
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: PC1, PKZ |
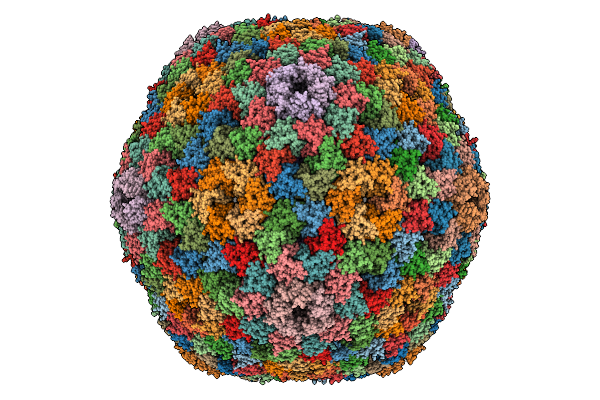 |
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
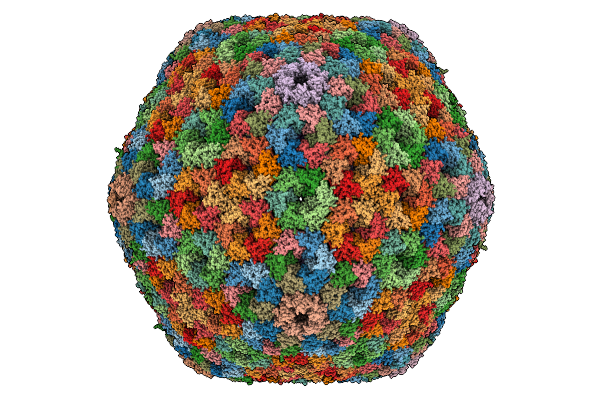 |
Cryo-Em Structure Of Carboxysomal Mid-Shell: T = 16 Shell Under C1 Symmetry.
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
 |
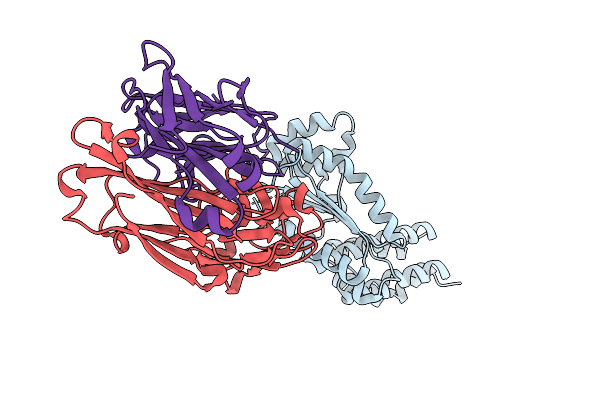
Crystal Structure Of Crimean-Congo Hemorrhagic Fever Virus Cap-Snatching Endonuclease
Organism: Orthonairovirus haemorrhagiae, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
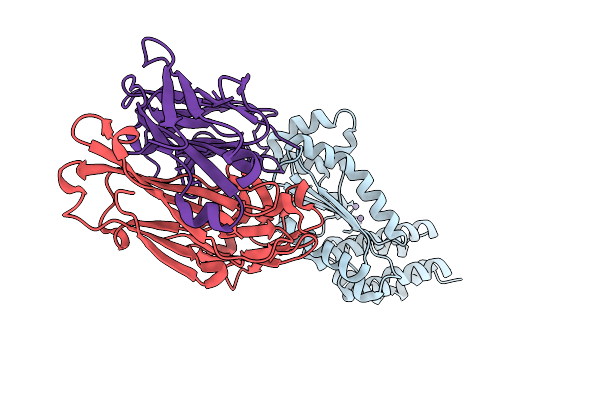
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
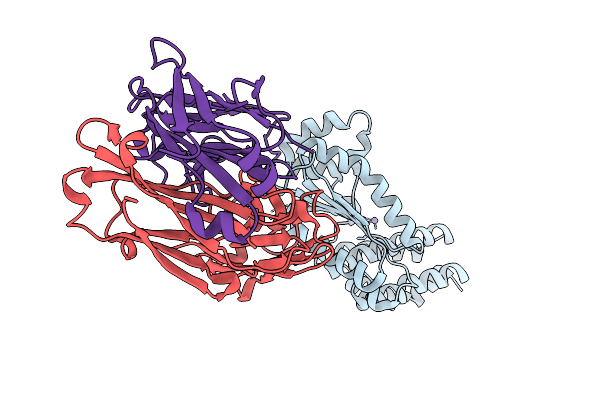
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Manganese Ions
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
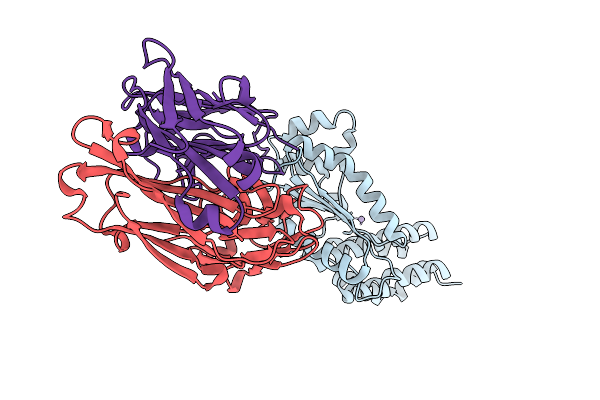
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E668A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
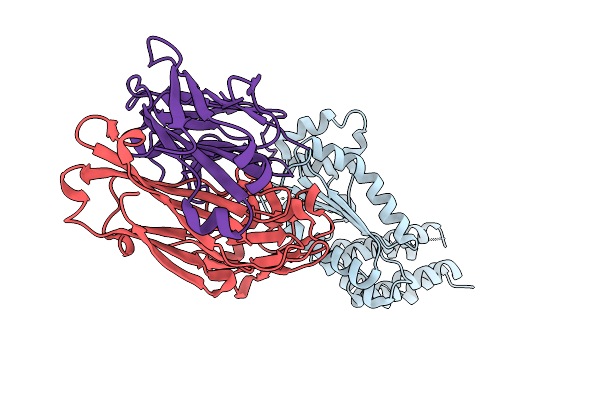
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E682A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (D719A Mutant)
Organism: Mus musculus, Kasokero virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With 2,4-Dioxo-4-Phenylbutanoic Acid (Dpba)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, XI7 |
 |
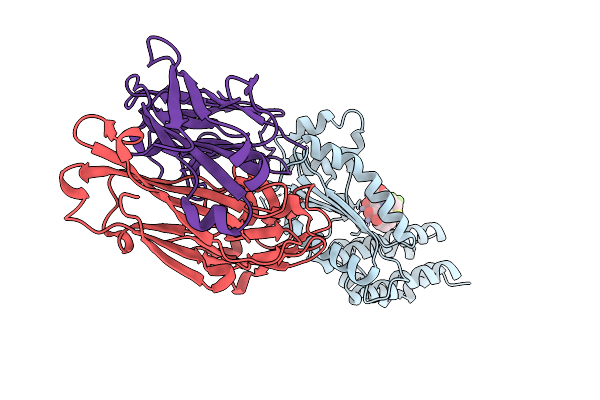
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With L-742,001
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, 0N8 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Baloxavir Acid (Bxa)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, E4Z |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: A1EH2 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: NIO |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |

