Search Count: 20
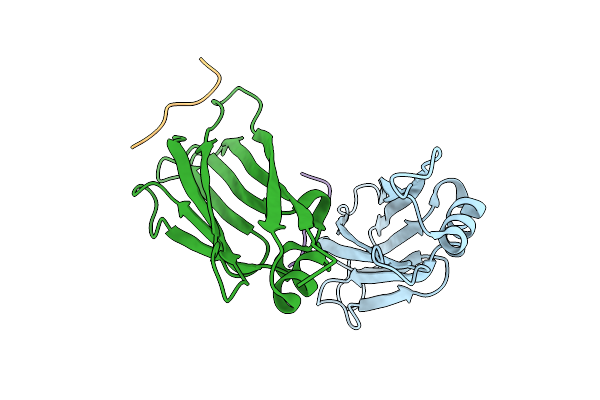 |
Crystal Structure Of Af9 Yeats Domain F28R Mutant In Complex With Histone H3K9La
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PEPTIDE BINDING PROTEIN Ligands: HOH |
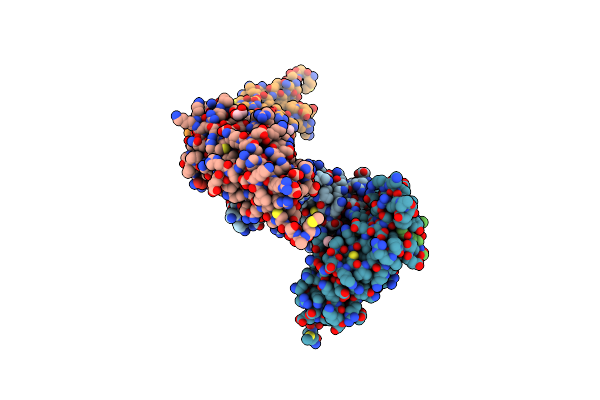 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2025-04-23 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO |
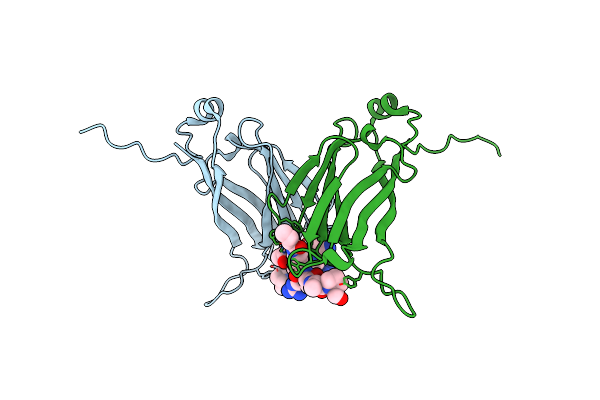 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-22 Classification: TRANSCRIPTION |
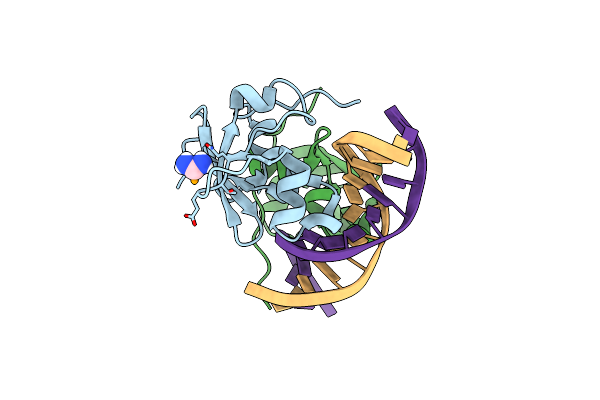 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: DNA BINDING PROTEIN/DNA Ligands: SEY |
 |
Selenomethionine Mutant (L601Sem/L654Sem) Of Sand Domain Of Protein Sp140 With Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: DNA BINDING PROTEIN/DNA |
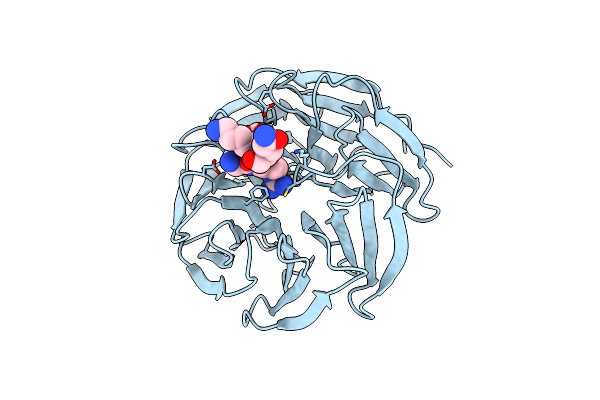 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-13 Classification: GENE REGULATION |
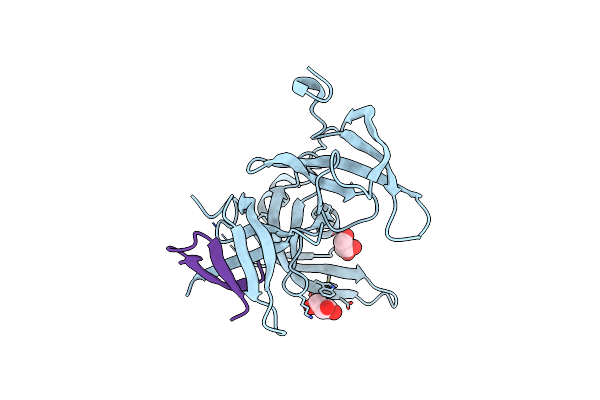 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: STRUCTURAL PROTEIN Ligands: GOL |
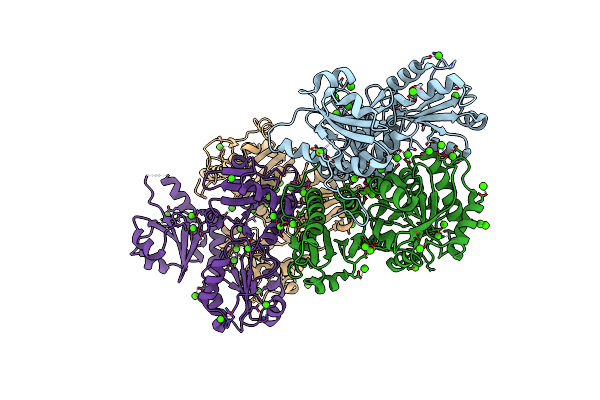 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-08 Classification: METAL BINDING PROTEIN Ligands: CA |
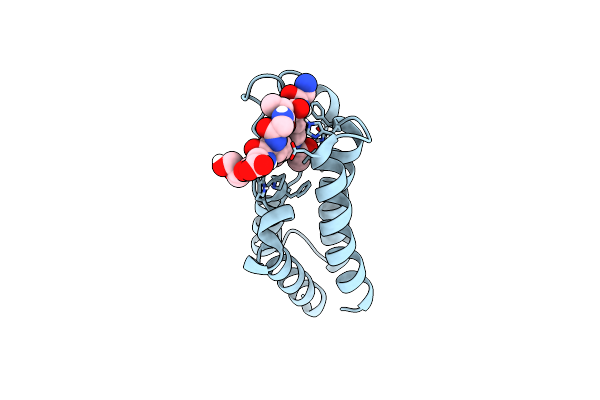 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-08-12 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2018-03-14 Classification: RNA BINDING PROTEIN Ligands: PO4 |
 |
Crystal Structure Of The Engineered Nine-Repeat Puf Domain In Complex With Cognate 9Nt-Rna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-03-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION/PEPTIDE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-04-20 Classification: SIGNALING PROTEIN/PEPTIDE Ligands: EDO, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION/PEPTIDE Ligands: CU, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-02-29 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure And Functional Study Of The Bowman-Birk Inhibitor From Rice Bran In Complex With Bovine Trypsin
Organism: Oryza sativa, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-07-22 Classification: Hydrolase inhibitor/Hydrolase |
 |
Crystal Structure Of C-Terminal Desundecapeptide Nitrite Reductase From Achromobacter Cycloclastes
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-12-20 Classification: OXIDOREDUCTASE Ligands: CU, CL |
 |
Crystal Structure Of Human Sulfotransferase Sult1A3 In Complex With Dopamine And 3-Phosphoadenosine 5-Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-08-30 Classification: TRANSFERASE Ligands: A3P, LDP |
 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph6.2
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, MES |
 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph5.0
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, ACY |

