Search Count: 22
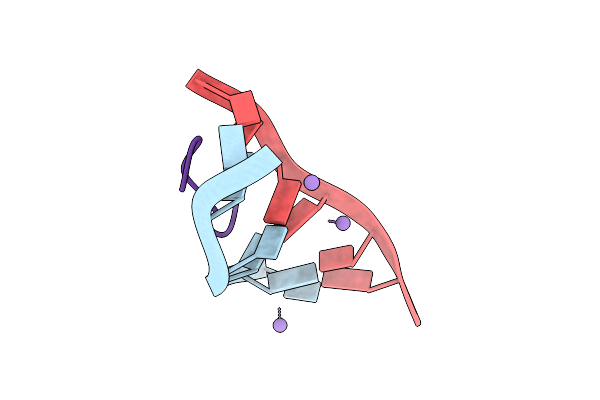 |
Crystal Structure Of Actinomycin D-Doxorubicin-D(Agccgt)2 Dna Ternary Complex
Organism: Synthetic construct, Streptomyces parvulus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-03-12 Classification: DNA/ANTIBIOTIC Ligands: DM2, MN, CL |
 |
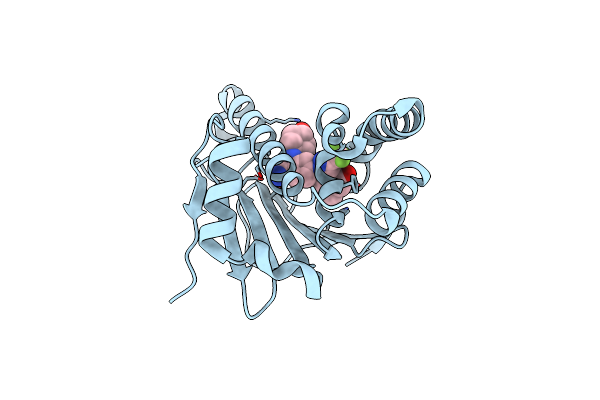
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-01-27 Classification: CHAPERONE Ligands: ET3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2021-01-27 Classification: CHAPERONE Ligands: E0G |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-01-13 Classification: CHAPERONE Ligands: EOR, GOL |
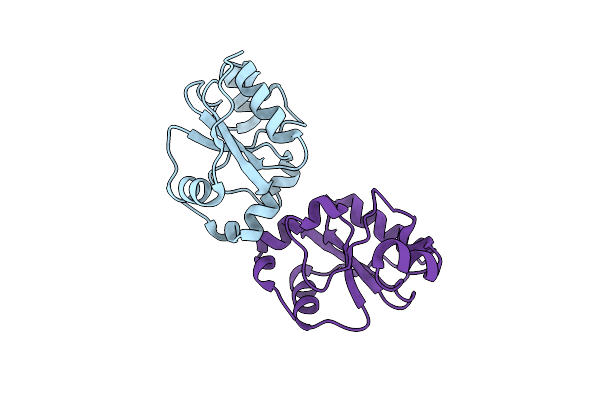 |
Crystal Structure Of Saccharomyces Cerevisiae Single Domain Sulfurtranferase Rdl2
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2019-11-06 Classification: TRANSFERASE |
 |
The Effect Of Isoleucine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Inhibition By Ligand Pt70 (Tcu)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
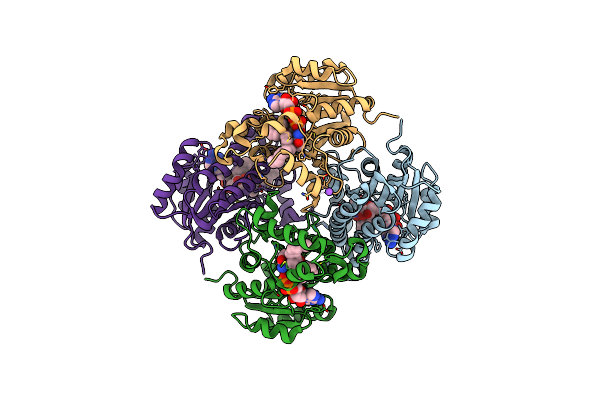
Compensation Of The Effect Of Isoleucine To Alanine Mutation By Designed Inhibition In The Inha Enzyme
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2015-08-12 Classification: OXIDOREDUCTASE Ligands: NAD, 53K |
 |
The Effect Of Valine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Substrate Binding Loop Conformation And Flexibility
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE Ligands: NAD, TCU, NA |
 |
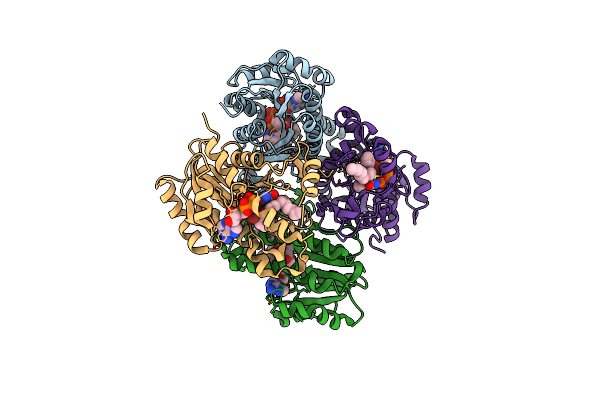
The Effect Of Isoleucine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Substrate Binding Loop Conformation And Flexibility
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE Ligands: NAD, TCU, ETE, EPE |
 |
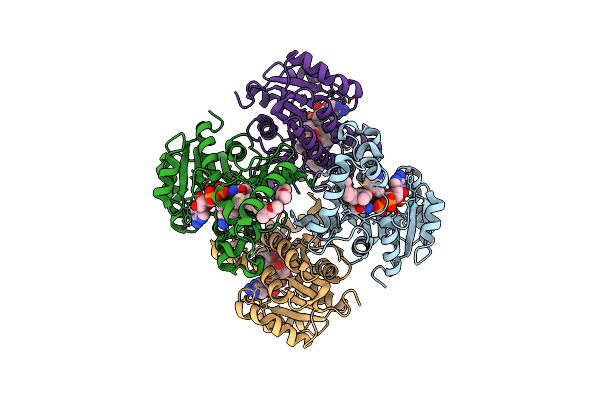
Crystal Structure Of Mycobacterium Tuberculosis Inha In Complex With Inhibitor Pt92
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 2TK |
 |
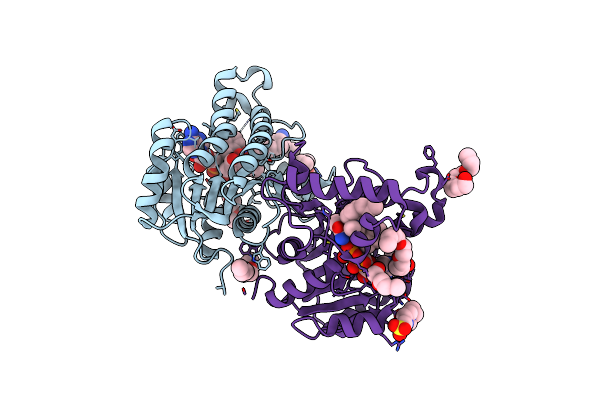
Multiple Binding Modes Of Inhibitor Pt155 To The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha Within A Tetramer
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-04-30 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: NAD, 1S5, 2NV |
 |
Substrate-Like Binding Mode Of Inhibitor Pt155 To The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1S5, CL, 2NV, EPE |
 |
Substrate-Binding Loop Movement With Inhibitor Pt10 In The Tetrameric Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1TN |
 |
Competition Of The Small Inhibitor Pt91 With Large Fatty Acyl Substrate Of The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha By Induced Substrate-Binding Loop Refolding
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1US |
 |
Crystal Structure Of Mycobacterium Tuberculosis Inha In Complex With Inhibitor Pt119 In 2.4 M Acetate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-04-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, JUS, ACT |
 |
Crystal Structure Of Escherichia Coli Menb In Complex With Substrate Analogue, Osb-Ncoa
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-24 Classification: LYASE/LYASE INHIBITOR Ligands: S0N, CL, PGE, GOL |
 |
Crystal Structure Of Escherichia Coli Menb, The 1,4-Dihydroxy-2-Naphthoyl-Coa Synthase In Vitamin K2 Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-08-24 Classification: LYASE Ligands: MLI, GOL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Menb In Complex With Substrate Analogue, Osb-Ncoa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-08-24 Classification: LYASE/LYASE INHIBITOR Ligands: S0N |
 |
Crystal Structure Of Mycobacterium Tuberculosis Menb With Altered Hexameric Assembly
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-08-24 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase (Inha) Inhibited By Triclosan
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-07 Classification: OXIDOREDUCTASE Ligands: TCL, NAD |

