Search Count: 24
 |
Cryo-Em Structure Of Formyl Peptide Receptor 2/C1R Receptor In Complex With Gi
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: STRUCTURAL PROTEIN Ligands: A1L1D |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: GOL, 1PE, OLC, RET |
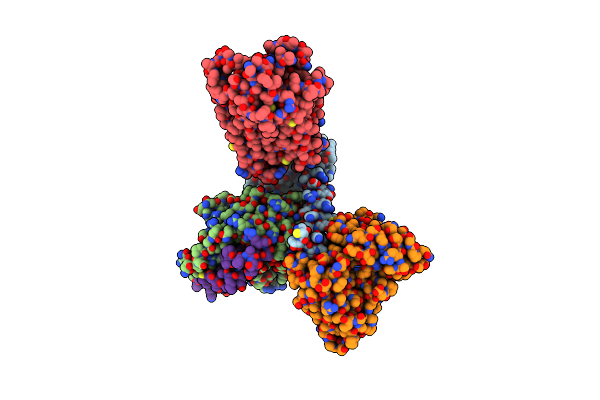 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: 7NR |
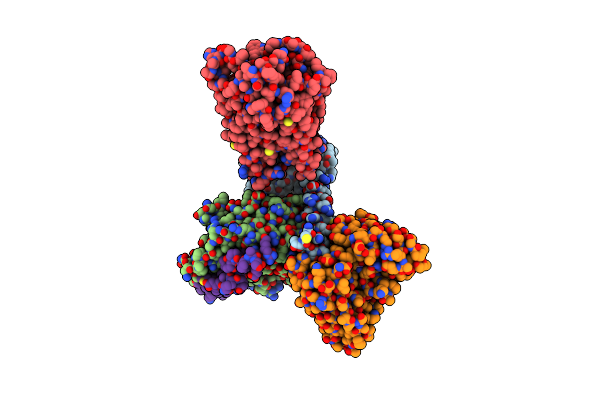 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: EIC |
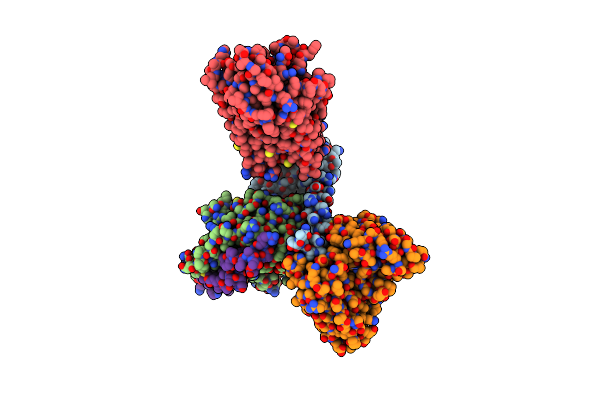 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: OLA |
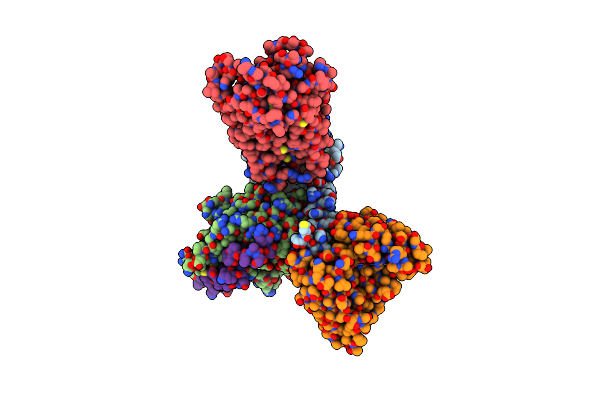 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: YN9 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: EPA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: MEMBRANE PROTEIN Ligands: YN9 |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P21221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, PEG, EDO, NA |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of C2221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, EDO, GOL, NA |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P1211
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, NA |
 |
Nmr Structure Of Foxo3A Transactivation Domains (Cr2C-Cr3) In Complex With Cbp Kix Domain (2B3L Conformation)
Organism: Mus musculus, Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-16 Classification: TRANSCRIPTION |
 |
Nmr Structure Of Foxo3A Transactivation Domains (Cr2C-Cr3) In Complex With Cbp Kix Domain (2L3B Conformation)
Organism: Mus musculus, Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-16 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-01-19 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Structure And Identification Of Adp-Ribose Recognition Motifs Of Aplf And Role In The Dna Damage Response
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-05-05 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-04-21 Classification: CELL ADHESION Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-04-21 Classification: CELL ADHESION |
 |
Structure Of E. Coli Toxin Rele (R81A/R83A) Mutant In Complex With Antitoxin Relbc (K47-L79) Peptide
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2009-03-17 Classification: Toxin/Toxin Repressor |

