Search Count: 22
 |
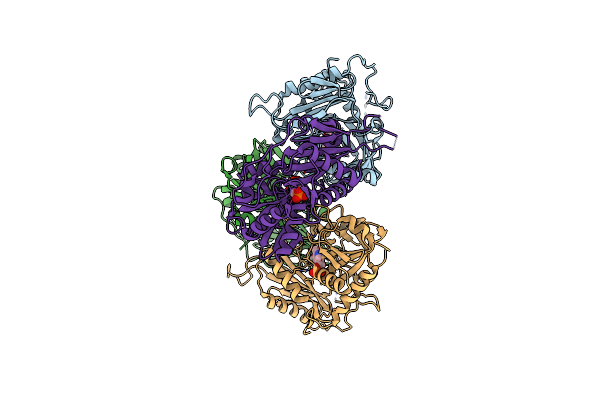
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde From The Cyanohydrin Cleavage
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: LYASE Ligands: NAG, FQ3, FAD, PGE, PEG, BCN |
 |
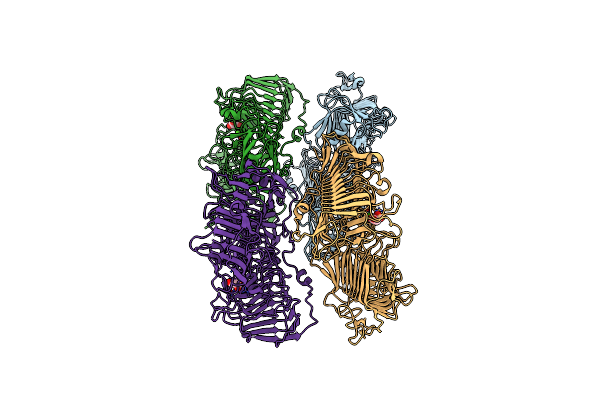
Organism: Rhodobacter sp. 140a
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-08 Classification: TRANSFERASE Ligands: PLP |
 |
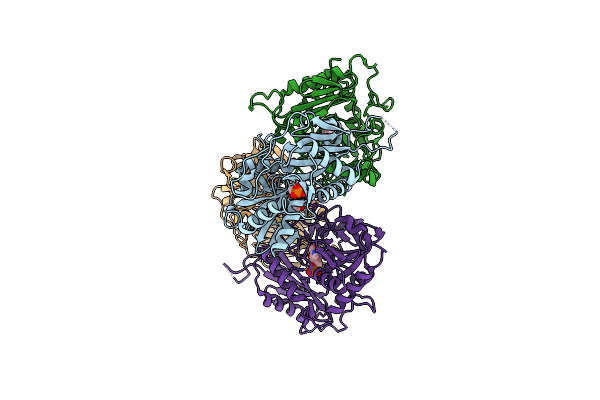
Organism: Bacteroides clarus
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-10-06 Classification: LYASE Ligands: CA, MLI |
 |
Organism: Rhodobacter sp. 140a
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PLP, EDO |
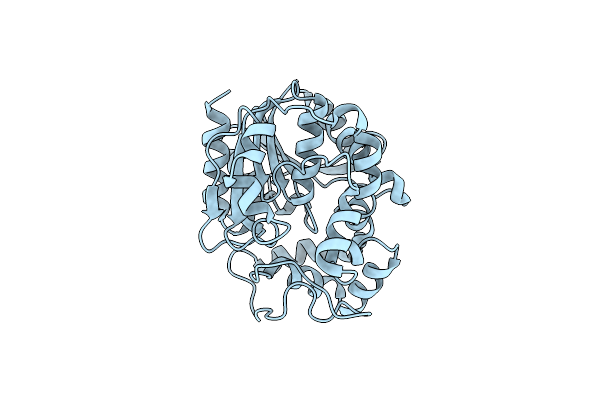 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-07 Classification: HYDROLASE |
 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-07 Classification: HYDROLASE |
 |
Crystal Complex Of Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Of Prunus Communis With Benzaldehyde
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-13 Classification: LYASE Ligands: NAG, FAD, HBX |
 |
Mutant L331A Of Deglycosylated Hydroxynitrile Lyase Isozyme 5 From Prunus Communis
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-13 Classification: LYASE Ligands: NAG, FAD |
 |
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-13 Classification: LYASE Ligands: MXN, NAG, FAD, PO4, PEG |
 |
Crystal Structure Of Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Of Prunus Communis
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-01 Classification: LYASE Ligands: NAG, FAD |
 |
Crystal Structure Of The Catalytic Domain Of A Multi-Domain Alginate Lyase Dp0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
Organism: Defluviitalea phaphyphila
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2019-10-30 Classification: LYASE Ligands: MN, MG, CA, ACT, EDO, CAC |
 |
Crystal Structure Of The Catalytic Domain Of A Multi-Domain Alginate Lyase Dp0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
Organism: Defluviitalea phaphyphila
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2019-10-30 Classification: LYASE Ligands: MN, CA, ACT, MG |
 |
Crystal Structure Of The Catalytic Domain Of A Multi-Domain Alginate Lyase Dp0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
Organism: Defluviitalea phaphyphila
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-10-30 Classification: LYASE Ligands: MN, CA, MG |
 |
Organism: Defluviitalea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-24 Classification: LYASE Ligands: SO4 |
 |
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE |
 |
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE |
 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-12-05 Classification: HYDROLASE |
 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-05 Classification: HYDROLASE |
 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-09-05 Classification: HYDROLASE |
 |
Organism: Vigna radiata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: SNO |

