Search Count: 13
 |
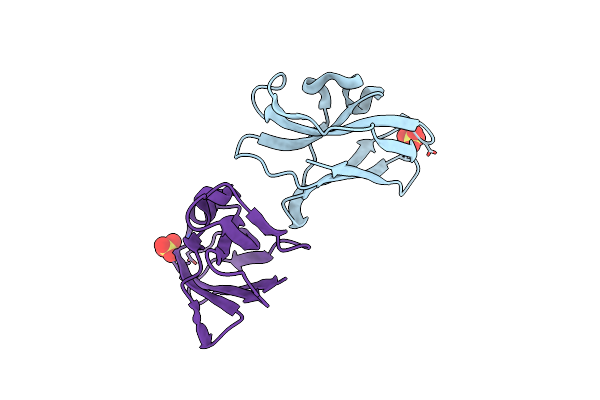
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: CL, ZQW |
 |
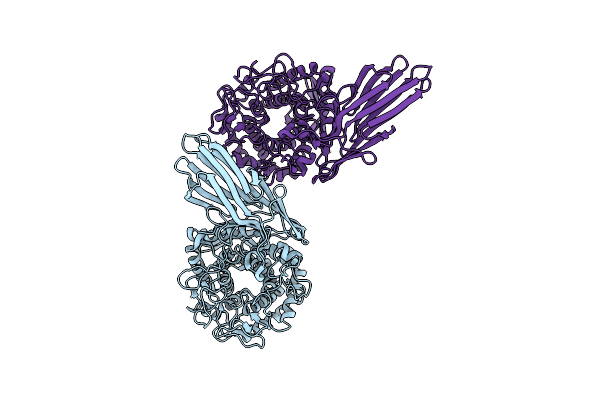
Co-Crystal Structure Of Capcna Bound To The Aoh1996 Derivative, Aoh1996-1Le
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.77 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ZQZ, CL |
 |
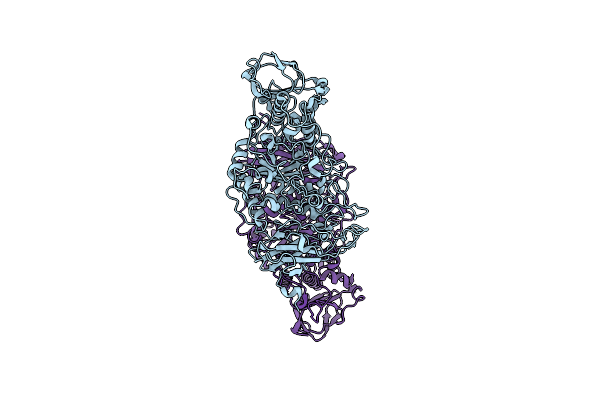
The X-Ray Crystallographic Structure Of Amylo-Alpha-1,6-Glucosidase From Thermococcus Gammatolerans Stb12
Organism: Thermococcus gammatolerans ej3
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-10-12 Classification: HYDROLASE |
 |
The X-Ray Crystallographic Structure Of Glycogen Debranching Enzyme From Sulfolobus Solfataricus Stb09
Organism: Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-03-09 Classification: HYDROLASE |
 |
The Crystal Structure Of Family 20 Cbm Of Maltotetraose-Forming Amylase From Pseudomonas Saccharophila Stb07
Organism: Pelomonas saccharophila
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: SO4 |
 |
 |
The Structure Of Maltooligosaccharide-Forming Amylase From Pseudomonas Saccharophila Stb07 With Maltotriose
Organism: Pelomonas saccharophila
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-01-15 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO |
 |
The Structure Of Maltooligosaccharide-Forming Amylase From Pseudomonas Saccharophila Stb07 With Maltotetraose
Organism: Pelomonas saccharophila
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-18 Classification: SUGAR BINDING PROTEIN Ligands: EDO, CA |
 |
Organism: Paenibacillus macerans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Giardia Guanine Phosphoribosyltransferase Complexed With A Transition State Analogue
Organism: Giardia intestinalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2000-07-26 Classification: TRANSFERASE Ligands: MG, IMU, POP, IPA |
 |
Crystal Structure Of Giardia Guanine Phosphoribosyltransferase Complexed With Immucilling
Organism: Giardia intestinalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2000-07-26 Classification: TRANSFERASE Ligands: IMG, IPA |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-08-18 Classification: TRANSFERASE Ligands: MG, IRP, POP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-06-22 Classification: PHOSPHORIBOSYLTRANSFERASE Ligands: MG, IMU, POP |

