Search Count: 1,641
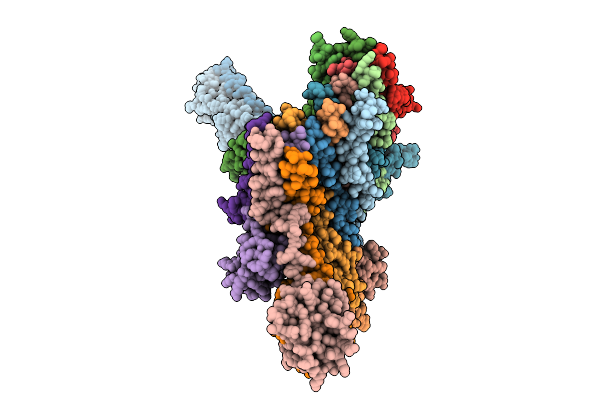 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Lacking The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
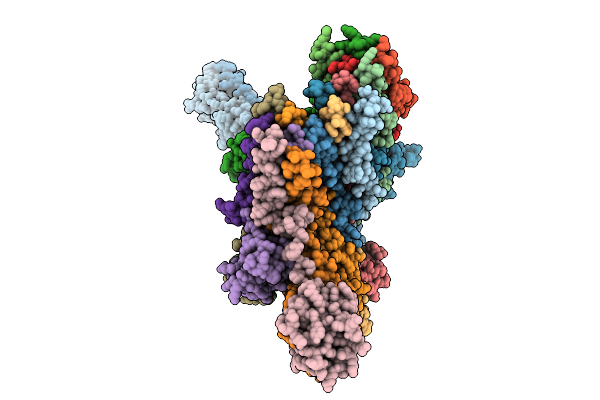 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Including The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
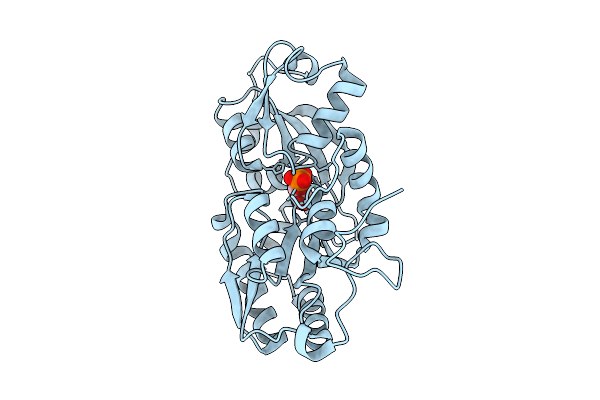 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
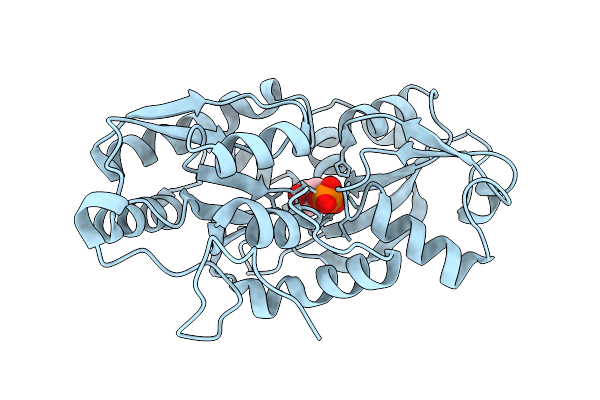 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: A1EG7 |
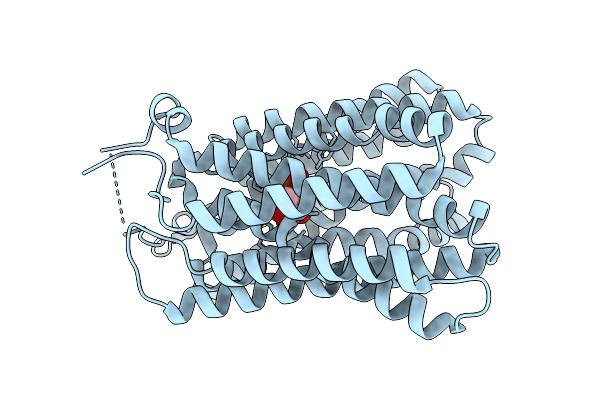 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: BG6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: RNA BINDING PROTEIN Ligands: GOL |
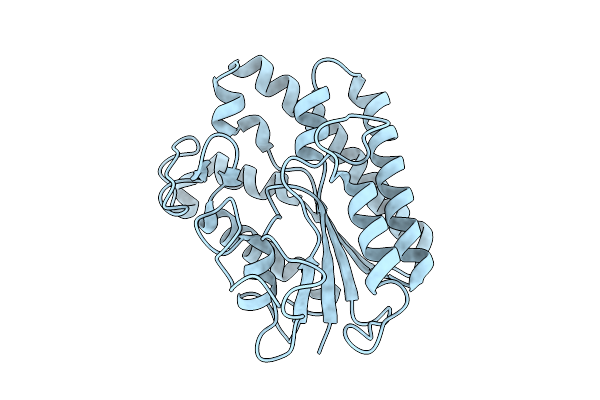 |
Crystal Structure Of Anae In Apo Form, A Fungal Gdsl-Acetylesterase With Acetylcholinesterase-Like Activity From Aspergillus Niger
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |
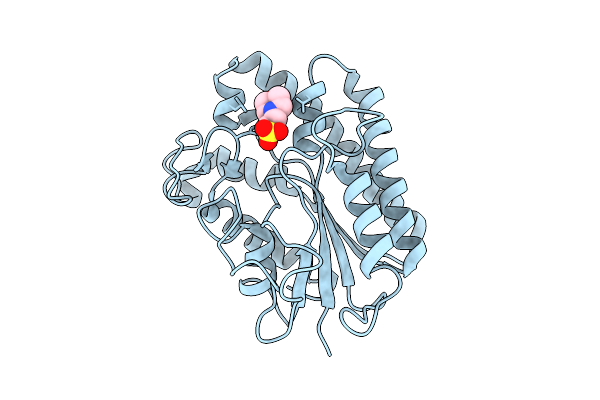 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: MES |
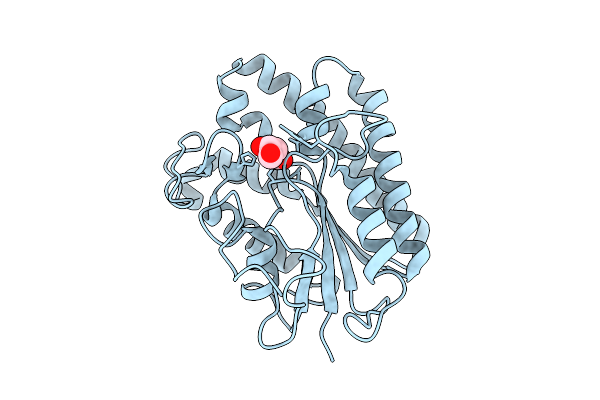 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: ACT, GOL |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: ACT, GOL |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: GOL, DPF |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: MES |

