Search Count: 383
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Fgd2 (Rv0132C) From Mycobacterium Tuberculosis Crystallised With Anderson-Evans Polyoxotungstate
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: TEW, IMD, MES |
 |
Fgd2 (Rv0132C) From Mycobacterium Tuberculosis With Cofactor F420 Crystallised With Anderson-Evans Polyoxotungstate
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: TEW, IMD, MPD, F42 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Cytochrome P450 Cyp125 In Complex With An Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, A1H47 |
 |
X-Ray Crystal Structure Of Cyp142 From Mycobacterium Tuberculosis In Complex With A Fragment Bound In Two Poses
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: 3QO, HEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, A1IGD, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD, PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, PEG, CL, PGE |
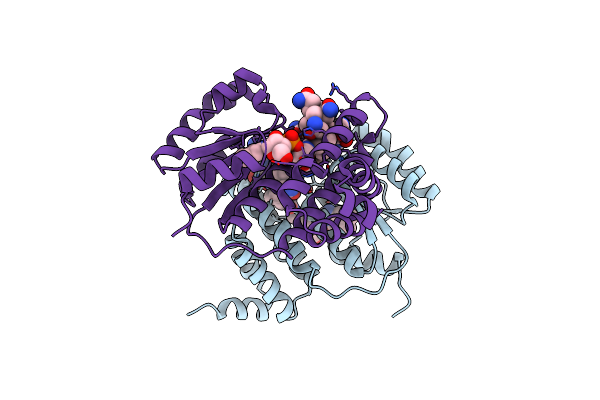 |
B12-Binding Domain From Chloracidobacterium Thermophilum Merr Family Protein, Dark State
Organism: Chloracidobacterium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: B12, 5AD |
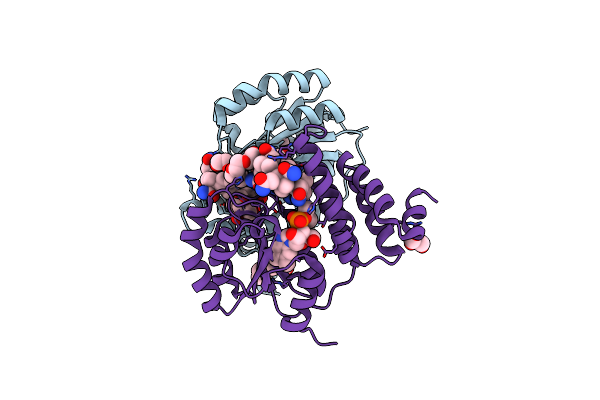 |
B12-Binding Domain From Chloracidobacterium Thermophilum Merr Family Protein, Anaerobic Light State
Organism: Chloracidobacterium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: B12, TRS, P6G, PG4, GOL |
 |
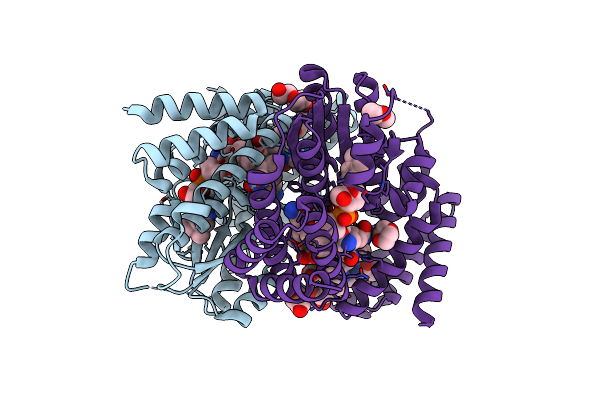
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-06-12 Classification: FLAVOPROTEIN Ligands: PO4 |
 |
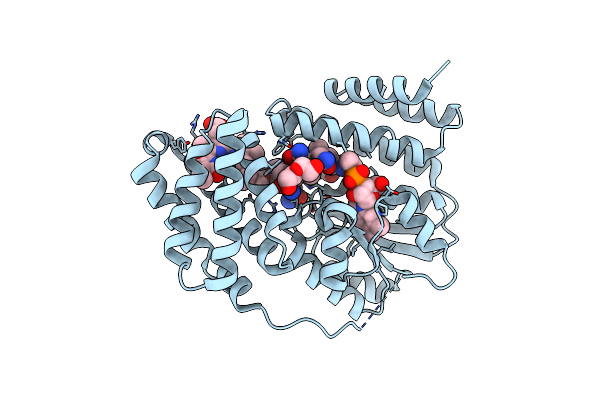
Organism: Saccharothrix syringae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-10 Classification: UNKNOWN FUNCTION Ligands: B12, 5AD, BLA, DIO, PEG |
 |
Organism: Saccharothrix syringae
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-04-10 Classification: UNKNOWN FUNCTION Ligands: B12, BLA, PEG |
 |
Organism: Acidimicrobiaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-04-10 Classification: UNKNOWN FUNCTION Ligands: B12, 5AD, PEG, SCN |
 |
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN, PO4 |
 |
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) T40C-S315C (Db1) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) S145C-P289C (Db2) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |

