Search Count: 7
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease Sa T95A Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: SO4, GOL, CAC |
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease Sa Y51F Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease S24A Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2013-08-14 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-12 Classification: CELL ADHESION |
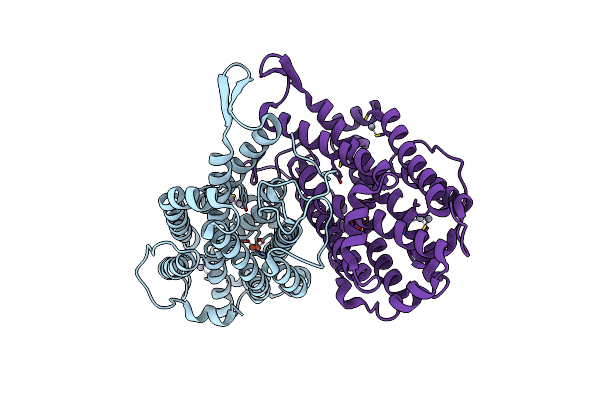 |
Dithionite Reduced E. Coli Ribonucleotide Reductase R2 Subunit, D84E Mutant
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-06-17 Classification: OXIDOREDUCTASE Ligands: FE, HG |
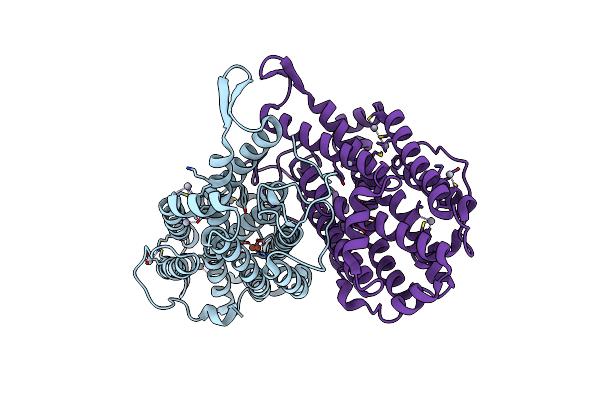 |
Oxidized Ribonucleotide Reductase R2-D84E Mutant Containing Oxo-Bridged Diferric Cluster
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-06-17 Classification: OXIDOREDUCTASE Ligands: FE, HG, O |
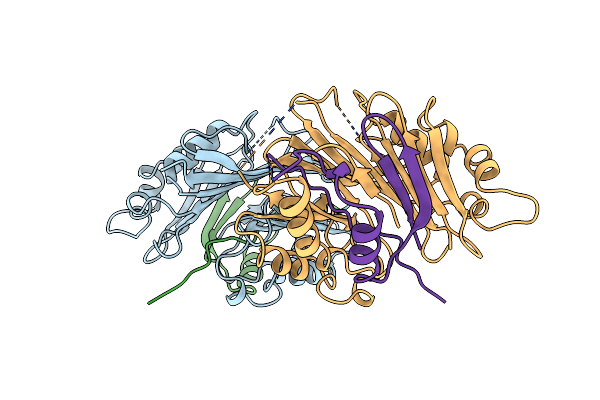 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 1999-06-01 Classification: S-ADENOSYLMETHIONINE DECARBOXYLASE |

