Search Count: 72
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
Crystal Structure Of Wild-Type Mycobacterium Tuberculosis Clpc1 N-Terminal Domain In Complex With Lassomycin
Organism: Mycobacterium tuberculosis, Lentzea kentuckyensis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-09-20 Classification: CHAPERONE Ligands: ACT |
 |
Crystal Structure Of Mycobacterium Tuberculosis R21K Clpc1 N-Terminal Domain In Complex With Lassomycin
Organism: Mycobacterium tuberculosis, Lentzea kentuckyensis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-09-20 Classification: CHAPERONE Ligands: ACT |
 |
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Erythromycin At 2.45A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-07-26 Classification: RIBOSOME Ligands: MG, HGR, ERY, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Azithromycin At 2.65A Resolution
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-07-26 Classification: RIBOSOME Ligands: MG, HGR, ZIT, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Telithromycin At 2.35A Resolution
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-07-26 Classification: RIBOSOME Ligands: MG, HGR, TEL, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Erythromycin At 2.50A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: RIBOSOME Ligands: MG, HGR, ERY, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Azithromycin At 2.50A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: RIBOSOME Ligands: MG, HGR, ZIT, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Telithromycin At 2.60A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-12 Classification: RIBOSOME Ligands: MG, HGR, TEL, MPD, ARG, ZN, SF4 |
 |
Organism: Photorhabdus australis
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.05 Å Release Date: 2022-10-19 Classification: ANTIBIOTIC |
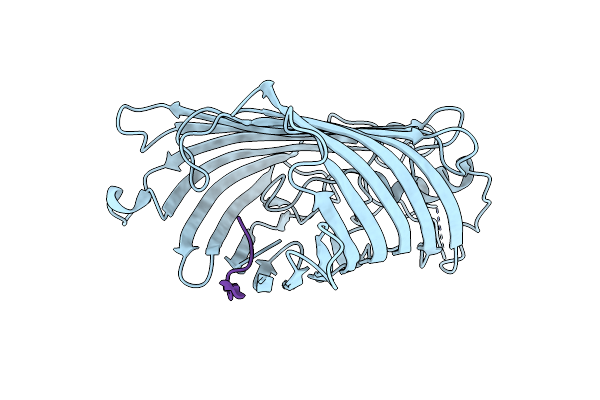 |
Organism: Escherichia coli o157:h7, Photorhabdus australis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN |
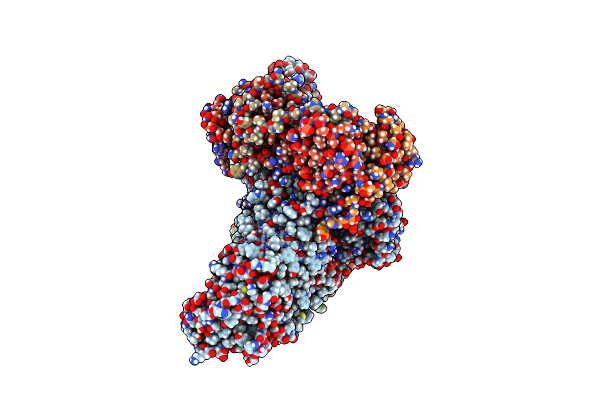 |
Organism: Escherichia coli k-12, Photorhabdus australis
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct, Photorhabdus noenieputensis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-08-17 Classification: ISOMERASE/ANTIBIOTIC/DNA Ligands: MG |
 |
Organism: Eleftheria terrae, staphylococcus aureus
Method: SOLID-STATE NMR Release Date: 2022-08-03 Classification: ANTIBIOTIC Ligands: 2PO, P1W |
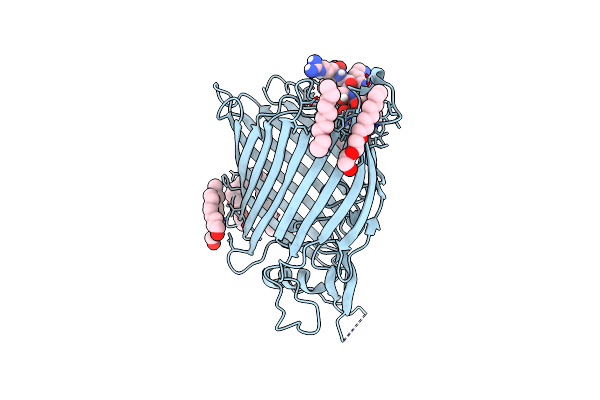 |
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: C8E |
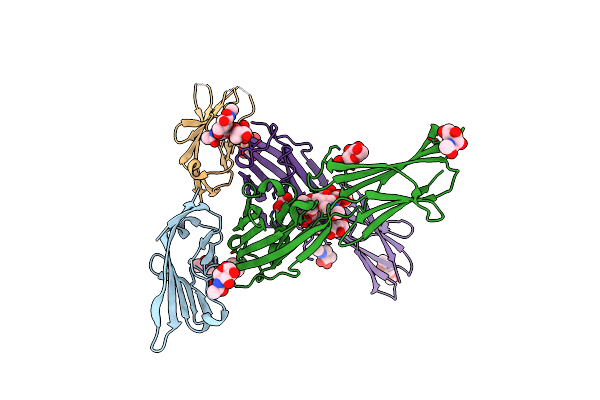 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2022-03-16 Classification: IMMUNE SYSTEM Ligands: NAG, GOL |
 |
Crystal Structure Of E.Coli Bama Beta-Barrel In Complex With Darobactin (Crystal Form 1)
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN Ligands: C8E, MG |
 |
Crystal Structure Of E.Coli Bama Beta-Barrel In Complex With Darobactin (Crystal Form 2)
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN Ligands: MG, C8E |

