Search Count: 12
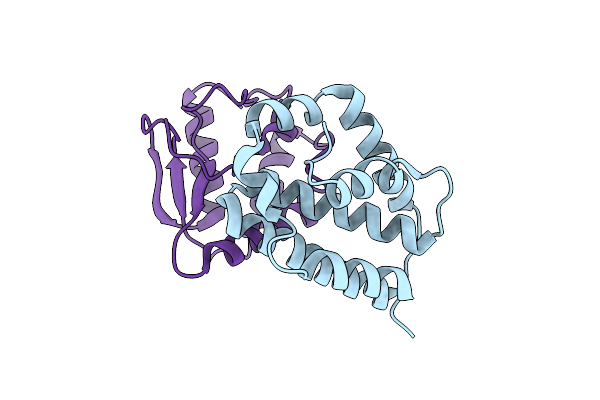 |
The Crystal Structure Of The Polymorphic Toxin Pt1(Em) H44A Mutant And Its Cognate Immunity Pim1(Em) Complex
Organism: Escherichia marmotae
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2024-07-03 Classification: TOXIN |
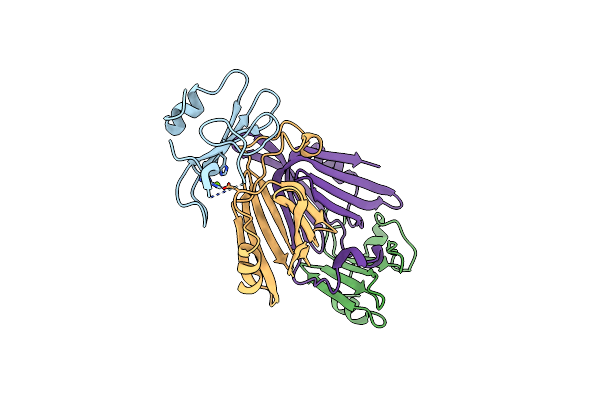 |
The Crystal Structure Of The Polymorphic Toxin Pt7(Bc) D37A Mutant And Its Cognate Immunity Pim7(Bc) Complex
Organism: Bacillus cereus bag3x2-1
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-06-19 Classification: TOXIN Ligands: MG |
 |
Crystal Structure Of Lugdulysin, A Staphylococcus Lugdunensis M30 Zinc Metallopeptidase
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: ZN, CA, CAC, ACT |
 |
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: CA, ZN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
 |
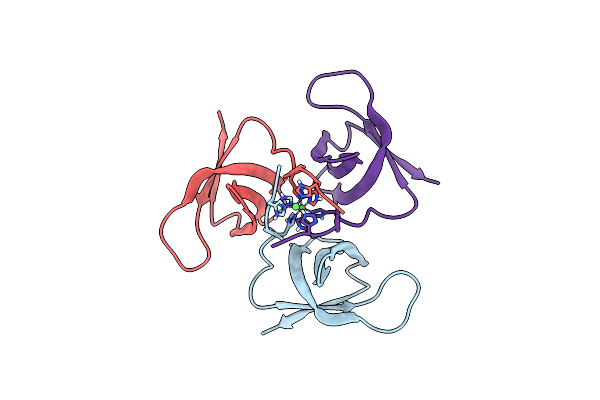
Crystal Structure Of The C-Terminal Domain Of The Hiv-1 Integrase (Subtype A2)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
 |
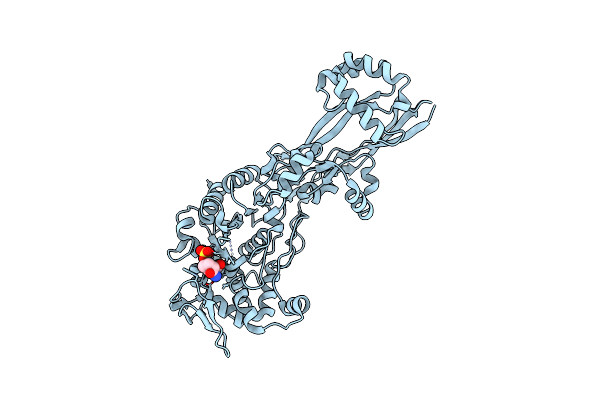
Crystal Structure Of The C-Terminal Domain Of The Hiv-1 Integrase (Subtype A2, Mutant N254K, K340Q)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
 |
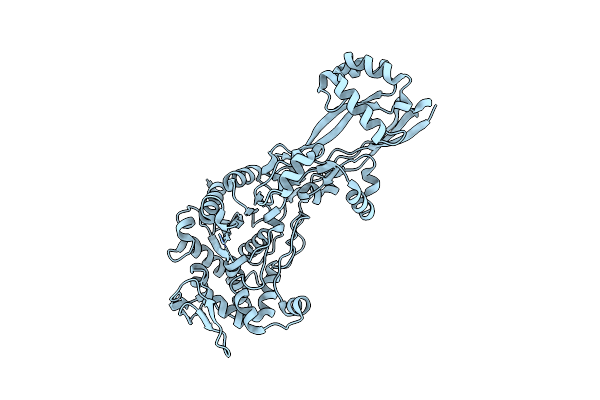
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC Ligands: NXL |
 |
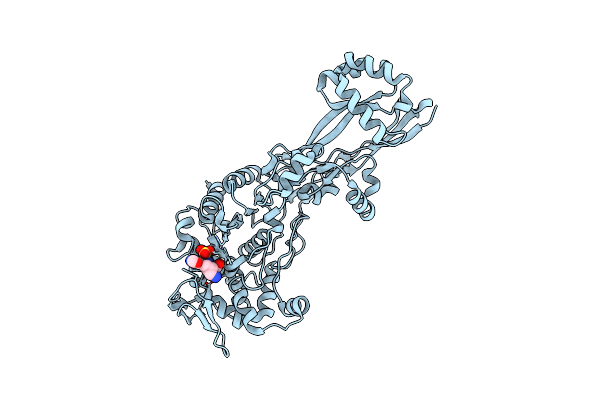
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC |
 |
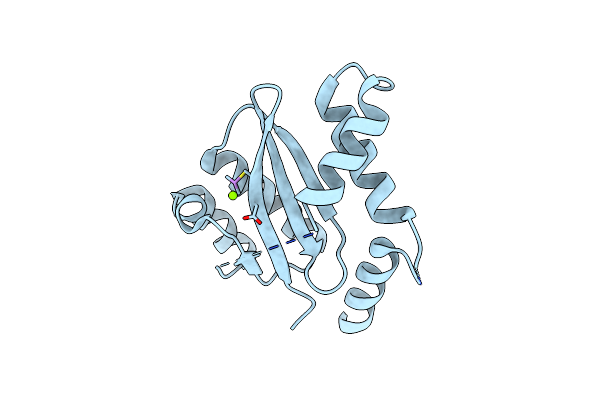
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC Ligands: ET5 |
 |
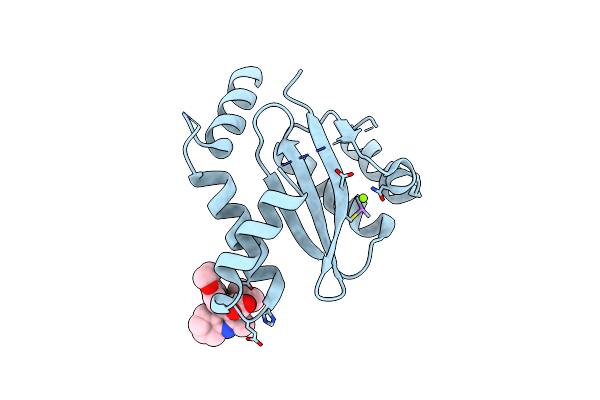
Dual Inhibition Of Hiv-1 Replication By Integrase-Ledgf Allosteric Inhibitors Is Predominant At Post-Integration Stage During Virus Production Rather Than At Integration
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: MG |
 |
Dual Inhibition Of Hiv-1 Replication By Integrase-Ledgf Allosteric Inhibitors Is Predominant At Post-Integration Stage During Virus Production Rather Than At Integration
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2013-12-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: LF0, MG |

