Search Count: 207
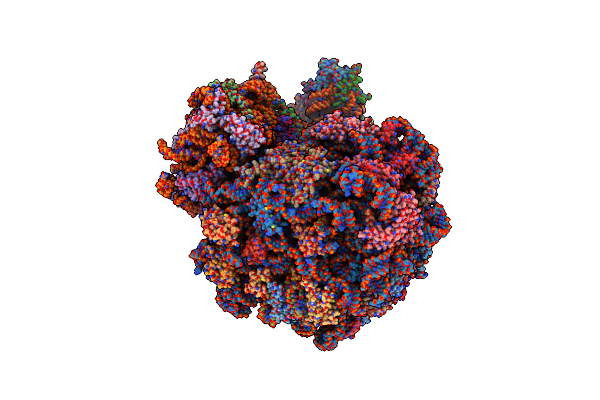 |
High-Resolution Cryo-Em Structure Of The Plasmodium Falciparum 80S Ribosome Bound To Rack1 And E-Trna
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RIBOSOME Ligands: MG, ZN |
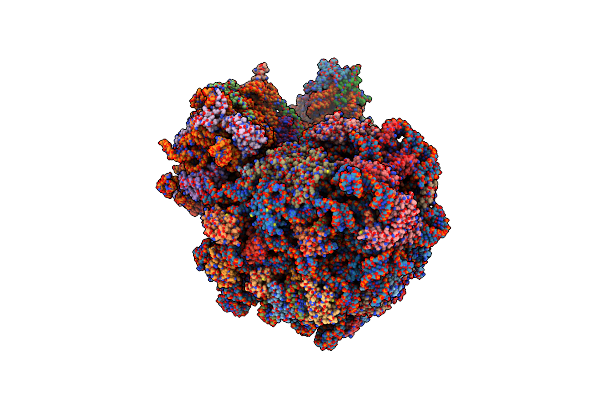 |
High-Resolution Cryo-Em Structure Of The Plasmodium Falciparum 80S Ribosome Bound To E-Trna
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RIBOSOME Ligands: MG, ZN |
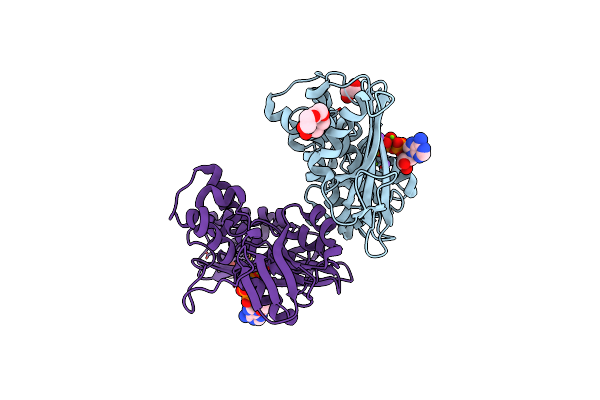 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-12 Classification: PROTEIN BINDING Ligands: MG, NA, ANP, GOL, PG4, EDO |
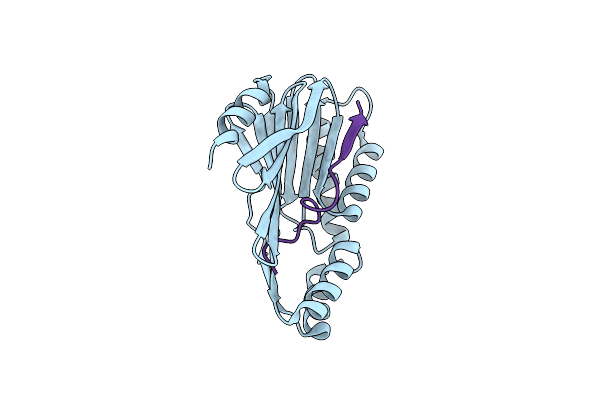 |
Connectase T1A C192S Mutant From Methanocaldococcus Mazei With Peptide Substrate
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-15 Classification: LIGASE |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-12-25 Classification: LIGASE |
 |
Organism: Plasmodium falciparum 3d7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: GOL, EDO, ZN, CL |
 |
Organism: Plasmodium falciparum 3d7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: PEG, ACY, ZN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: HYDROLASE Ligands: H8F, ZN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-08-07 Classification: PROTEIN BINDING Ligands: ACT, CA, TPS, ANP, PG4, PEG, EDO |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-01-17 Classification: TRANSFERASE/INHIBITOR Ligands: XUH, EDO, NA, PO4 |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: XUH, EDO, ZN |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: XUN, GOL, EDO, PEG, ZN |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-10-19 Classification: PROTEIN BINDING Ligands: 7TU, ACT, ZN, CL, NA |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Ny-Hydroxyasparagine: A Multifunctional Unnatural Amino Acid That Is A Good P1 Substrate Of Asparaginyl Peptide Ligases
Organism: Macaca mulatta
Method: SOLUTION NMR Release Date: 2022-09-07 Classification: METAL BINDING PROTEIN |
 |
The Crystal Structure Of Vypal2-C214A, A Dead Mutant Of Vypal2 Peptide Asparaginyl Ligase In Form Ii
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: PLANT PROTEIN Ligands: NAG |
 |
The Crystal Structure Of Vypal2-I244V, A More Efficient Mutant Of Vypal2 Peptide Asparaginyl Ligase In Its Active Enzyme Form
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN Ligands: NAG, EDO, GOL |

