Search Count: 47
 |
Crystal Structure Of The Native Copper Efflux Oxidase (Cueo) From Hafnia Alvei
Organism: Hafnia alvei
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: METAL BINDING PROTEIN Ligands: CU, EDO |
 |
Crystal Structure Of The Copper Efflux Oxidase (Cueo) From Hafnia Alvei Deleted Of The Met-Rich Domain
Organism: Hafnia alvei
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: METAL BINDING PROTEIN Ligands: CU, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-04-10 Classification: FLAVOPROTEIN Ligands: FAD, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-04-10 Classification: FLAVOPROTEIN |
 |
Organism: Flavobacterium johnsoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-03 Classification: UNKNOWN FUNCTION Ligands: MG, ZN, ACT |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Reduced Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL, ACT |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Oxidized Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, NA, CL, GOL |
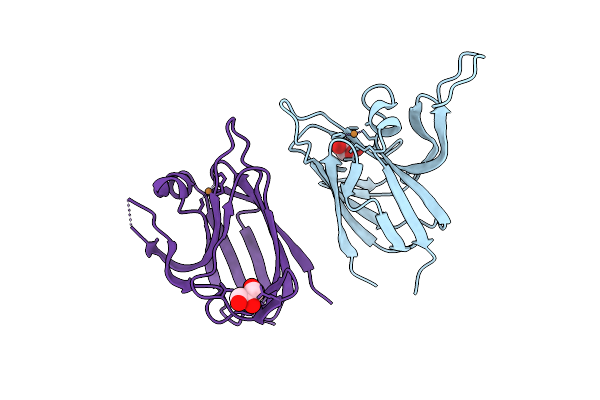 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, H166A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL |
 |
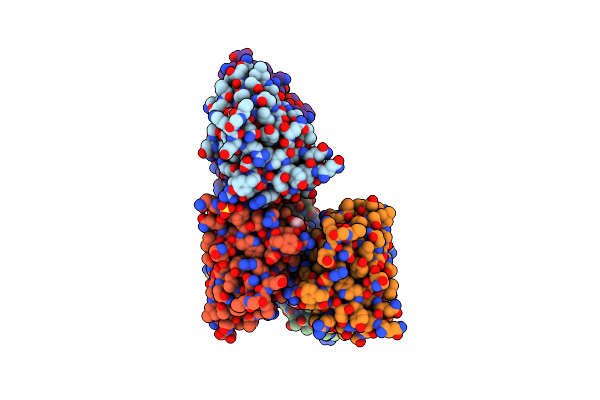
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, M171A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, GOL |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-01 Classification: IMMUNE SYSTEM Ligands: SO4, GOL, CXS |
 |
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-18 Classification: PROTEIN TRANSPORT |
 |
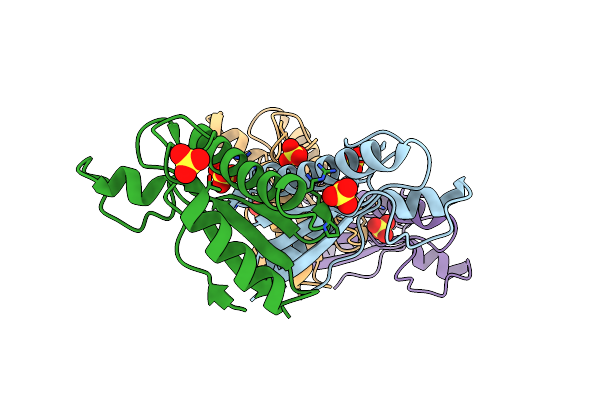
Organism: Drechmeria coniospora
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-19 Classification: IMMUNE SYSTEM Ligands: CL |
 |
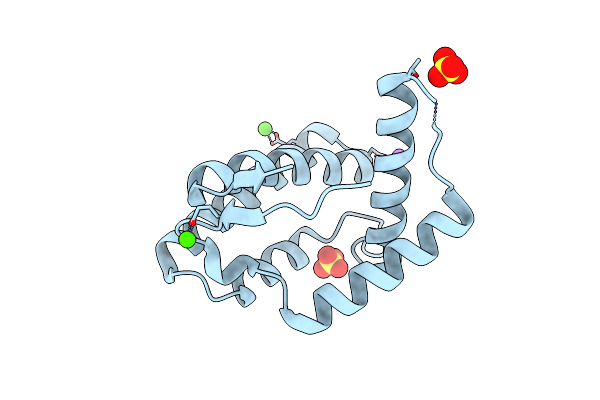
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-08-04 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Varroa destructor
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-07-07 Classification: TRANSPORT PROTEIN Ligands: SO4, CA, NA |
 |
Organism: Varroa destructor
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-07-07 Classification: TRANSPORT PROTEIN Ligands: SO4, NHE, NA |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-26 Classification: IMMUNE SYSTEM |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-05-26 Classification: IMMUNE SYSTEM |
 |
Structure Of The Pore C-Terminal Domain, A Protein Of T9Ss From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis jcvi sc001
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-06-17 Classification: PROTEIN BINDING Ligands: MLD, NA |
 |
Organism: Porphyromonas gingivalis, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-02-07 Classification: PROTEIN TRANSPORT |
 |
Organism: Flavobacterium johnsoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-02-07 Classification: PROTEIN TRANSPORT Ligands: EDO |

