Search Count: 9
 |
Organism: Candida auris
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-04-01 Classification: ISOMERASE Ligands: SO4 |
 |
Allosteric Coupling Of Conformational Transitions In The Fk1 Domain Of Fkbp51 Near The Site Of Steroid Receptor Interaction
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-05-13 Classification: ISOMERASE |
 |
Structural Basis Of Conformational Transitions In The Active Site And 80 S Loop In The Fk506 Binding Protein Fkbp12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-02-12 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-15 Classification: ISOMERASE Ligands: MLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-15 Classification: ISOMERASE |
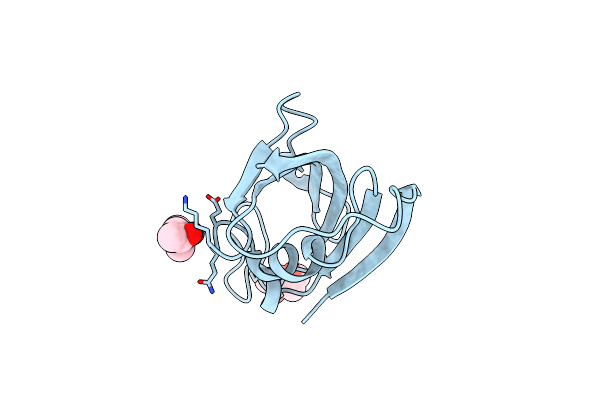 |
Analyzing The Visible Conformational Substates Of The Fk506 Binding Protein Fkbp12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-06-05 Classification: ISOMERASE Ligands: MPD |
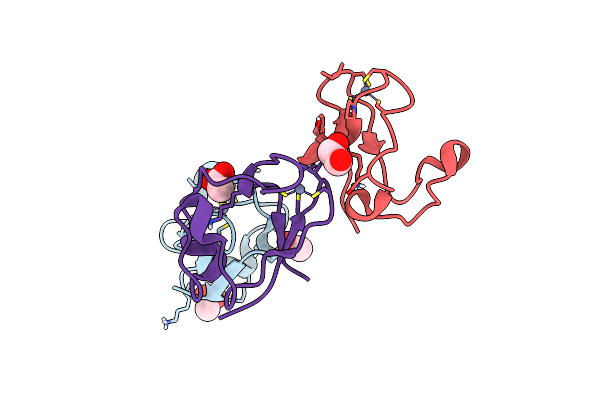 |
Nmr And X-Ray Analysis Of Structural Additivity In Metal Binding Site-Swapped Hybrids Of Rubredoxin
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:0.79 Å Release Date: 2007-12-18 Classification: ELECTRON TRANSPORT Ligands: ZN, ACT, EDO |
 |
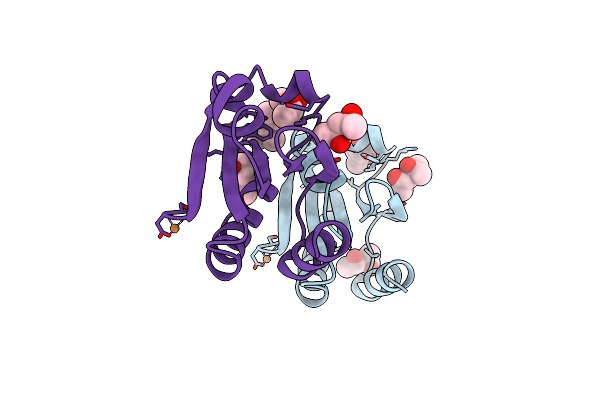
Nmr And X-Ray Analysis Of Structural Additivity In Metal Binding Site-Swapped Hybrids Of Rubredoxin
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2007-12-18 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Crystal Structure Of Thioredoxin From Escherichia Coli At 1.68 Angstroms Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 1991-10-15 Classification: ELECTRON TRANSPORT Ligands: CU, MPD |

