Search Count: 22
 |
Organism: Microbacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-10-06 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Microbacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2021-10-06 Classification: METAL BINDING PROTEIN Ligands: ZN, U1 |
 |
Crystal Structure Of A Selenomethionine-Substituted A70M I84M Mutant Of The Essential Repressor Ddro From Radiation Resistant-Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti (strain vcd115 / dsm 17065 / lmg 22923)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Crystal Structure Of The N-Terminal Hth Dna-Binding Domain Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The C-Terminal Dimerization Domain Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Minor Ferredoxin From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2019-09-18 Classification: ELECTRON TRANSPORT Ligands: SO4, FES |
 |
Organism: Thermosynechococcus elongatus bp-1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-09-18 Classification: ELECTRON TRANSPORT Ligands: BEN, FES, SO4 |
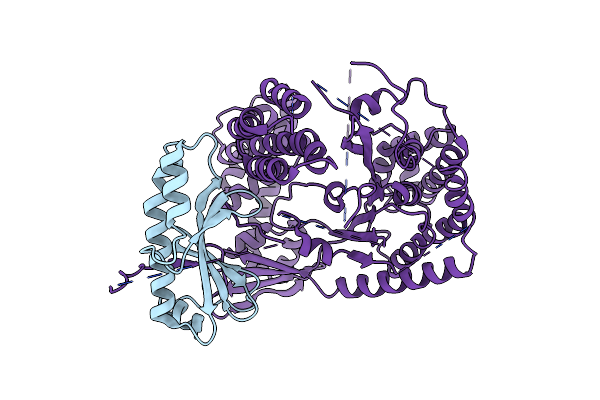 |
Structure Of The Heterodimeric Complex Exou-Spcu From The Type Iii Secretion System (T3Ss) Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2012-04-18 Classification: TRANSPORT PROTEIN |
 |
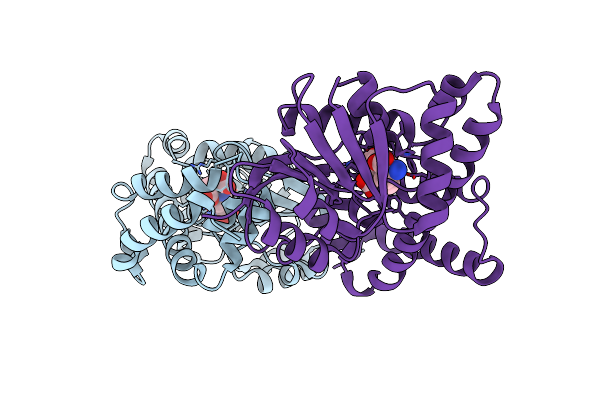
Structural Insights Into The Catalytic Mechanism And The Role Of Streptococcus Pneumoniae Pbp1B
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-05-26 Classification: TRANSFERASE Ligands: SO4, S2D, CL |
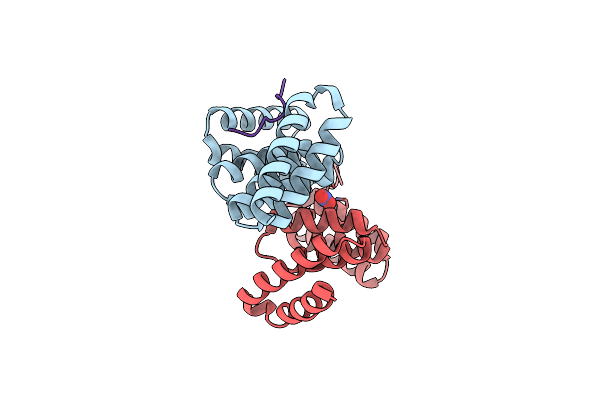 |
Crystal Structure Of Pcrh In Complex With The Chaperone Binding Region Of Popd
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-05-05 Classification: PROTEIN BINDING Ligands: NO3 |
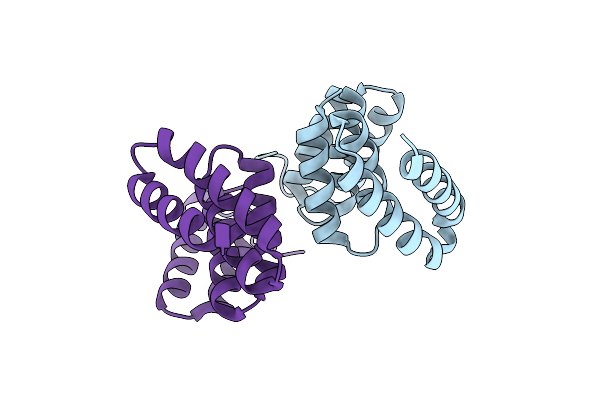 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2010-05-05 Classification: PROTEIN BINDING |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: TNA, BR |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: TNA, MTA, BR |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: BR, B3P |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: GLU, NA |
 |
Crystal Structure Of E81Q Mutant Of Mtnas In Complex With S-Adenosylmethionine
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: SAM, BR, B3P |
 |
Structural Characterization Of A Putative Endogenous Metal Chelator In The Periplasmic Nickel Transporter Nika (Nickel Butane-1,2,4-Tricarboxylate Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-09-16 Classification: METAL TRANSPORT Ligands: NI, ACT, SO4, CL, HCT, GOL |
 |
Structural Characterization Of A Putative Endogenous Metal Chelator In The Periplasmic Nickel Transporter Nika (Butane-1,2,4-Tricarboxylate Without Nickel Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-09-16 Classification: METAL TRANSPORT Ligands: GOL, ACT, SO4, HCT, CL |
 |
Crystallographic And Spectroscopic Evidence For High Affinity Binding Of Fe Edta (H2O)- To The Periplasmic Nickel Transporter Nika
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-08-02 Classification: HYDROLASE Ligands: FE, ACT, SO4, CL, EDT, DTT, GOL |

