Search Count: 16
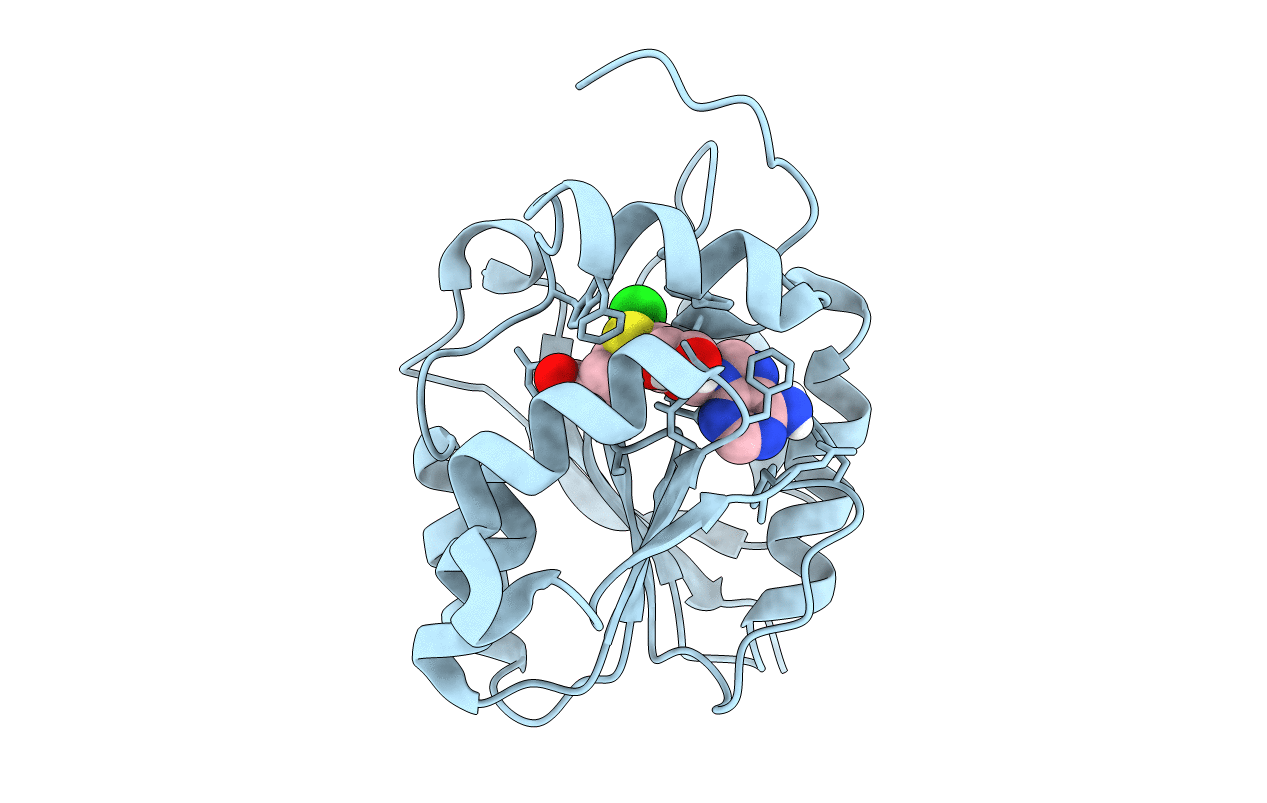 |
Crystal Structure Of Halogen Methyl Transferase From Paraburkholderia Xenovorans At 1.8 A In Complex With Sah
Organism: Paraburkholderia xenovorans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: SAH, CL |
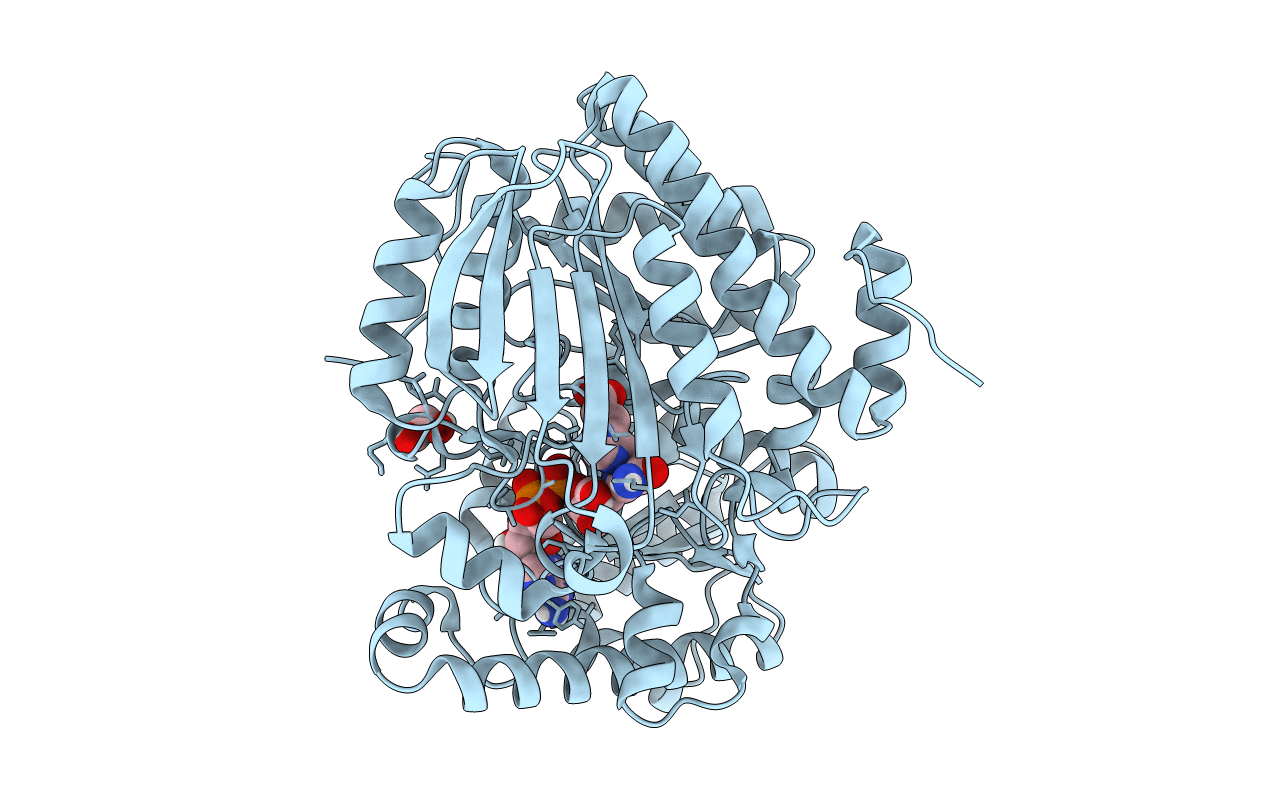 |
Thiourocanate Hydratase From Paenibacillus Sp. Soil724D2 In Complex With Cofactor Nad+ And Urocanate
Organism: Paenibacillus sp. soil724d2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-02-24 Classification: LYASE Ligands: URO, NAD, EDO |
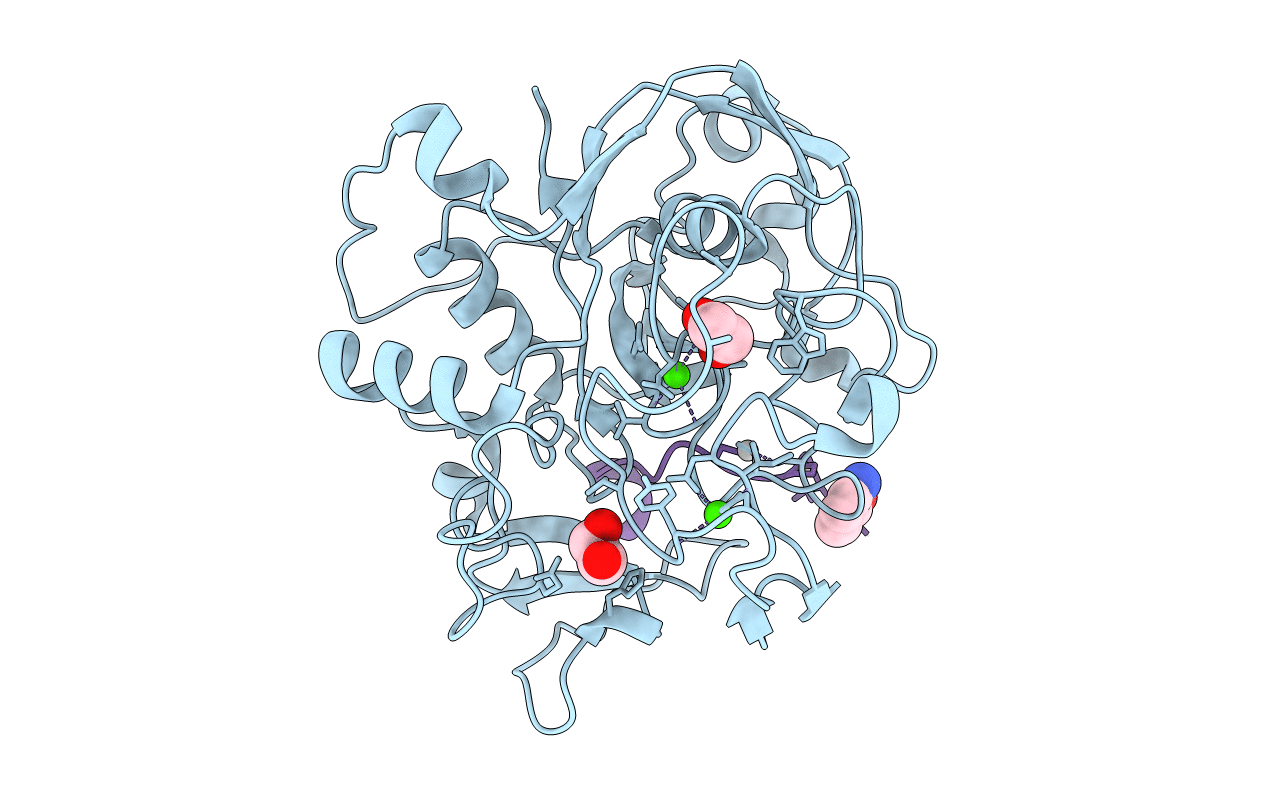 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: AG, CA, EDO, CL, SOA |
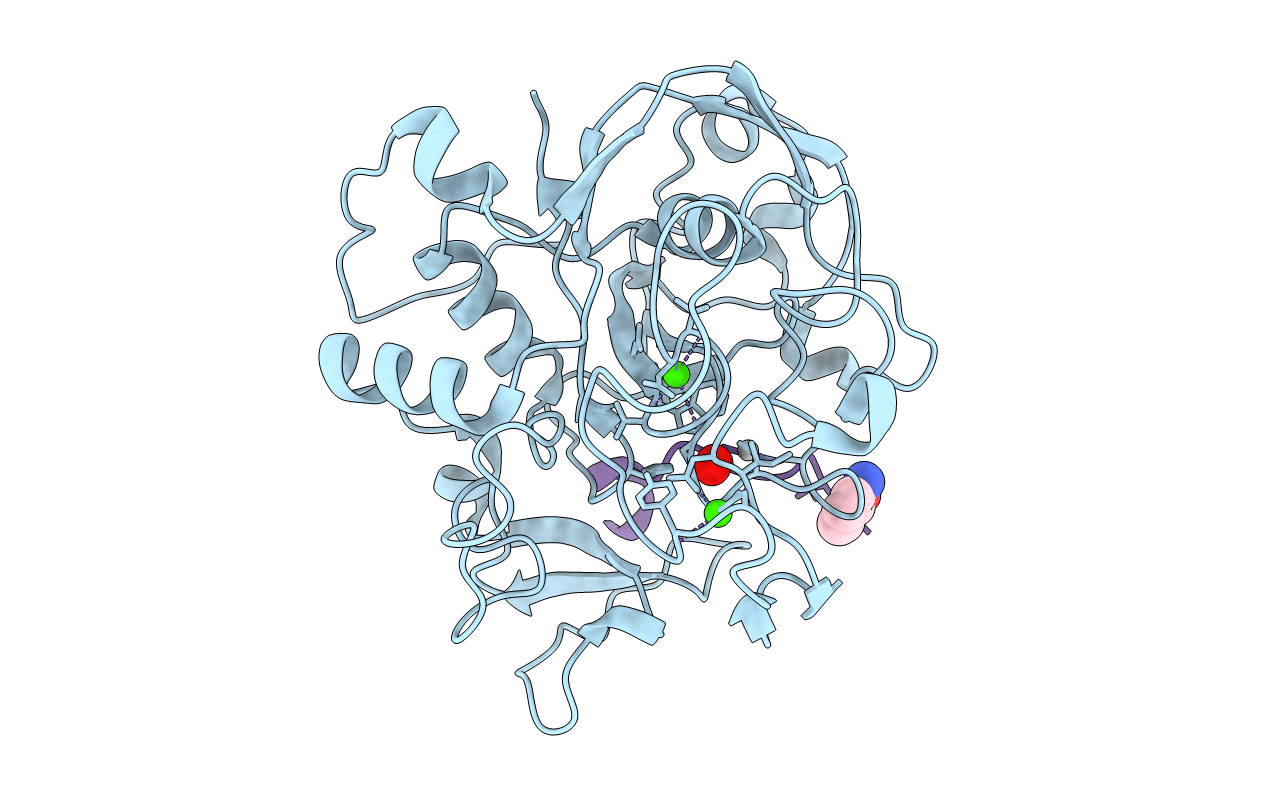 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S:O2 Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: OXY, CA, CL, AG, SOA |
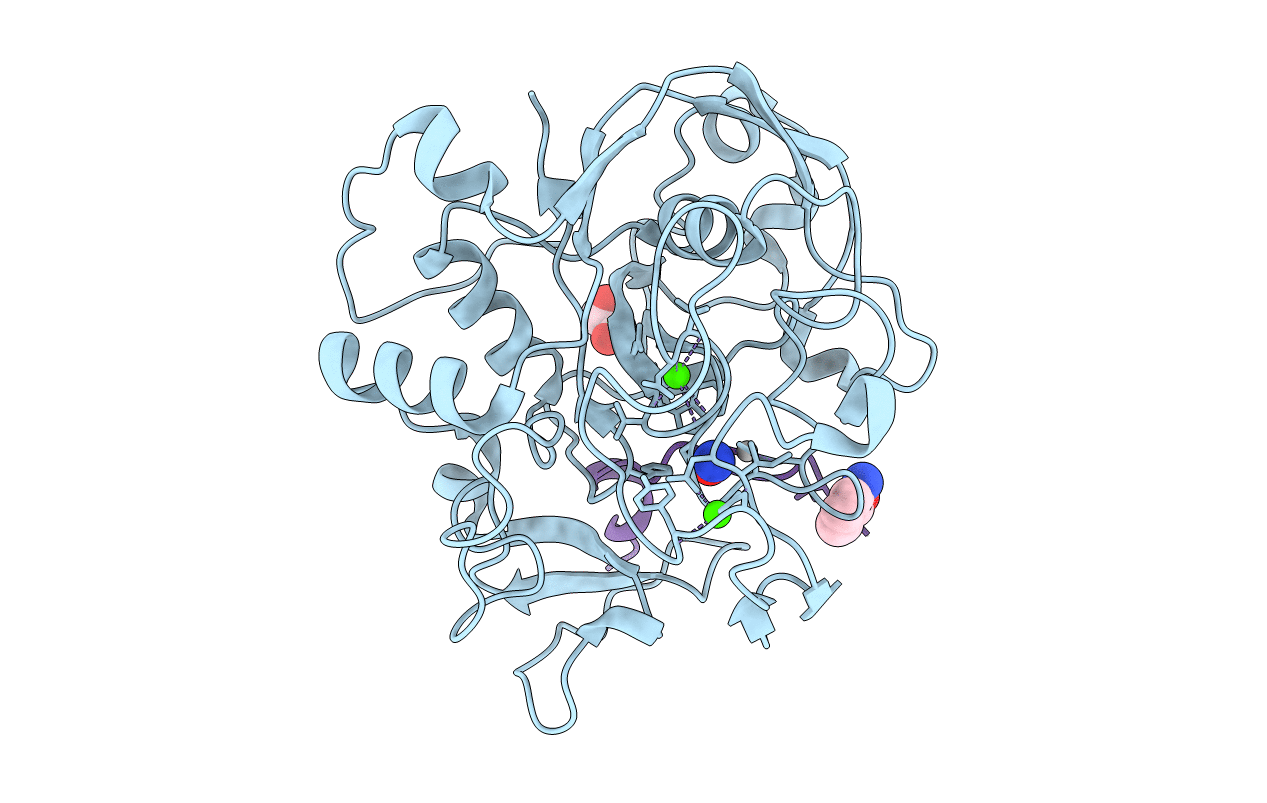 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S:No Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: AG, CA, CL, NO, EDO, SOA |
 |
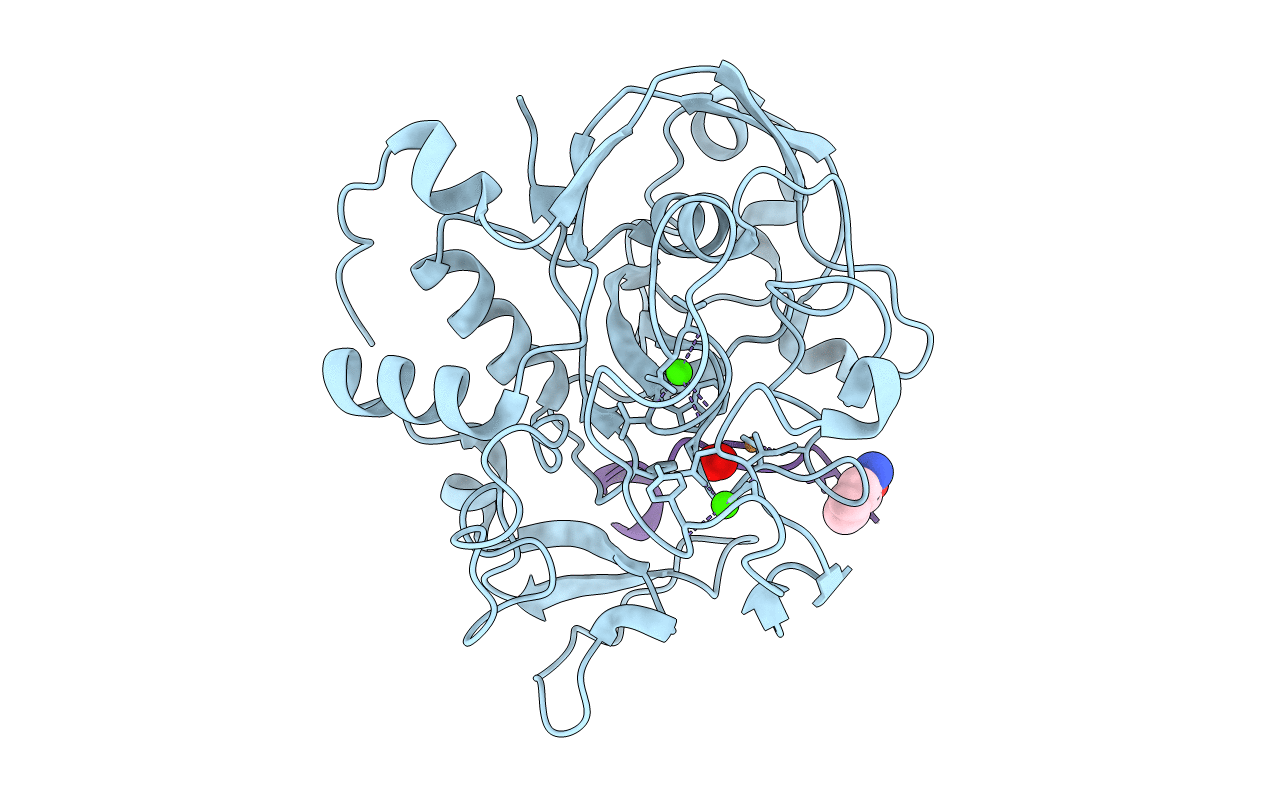
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:No Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, NO, SOA |
 |
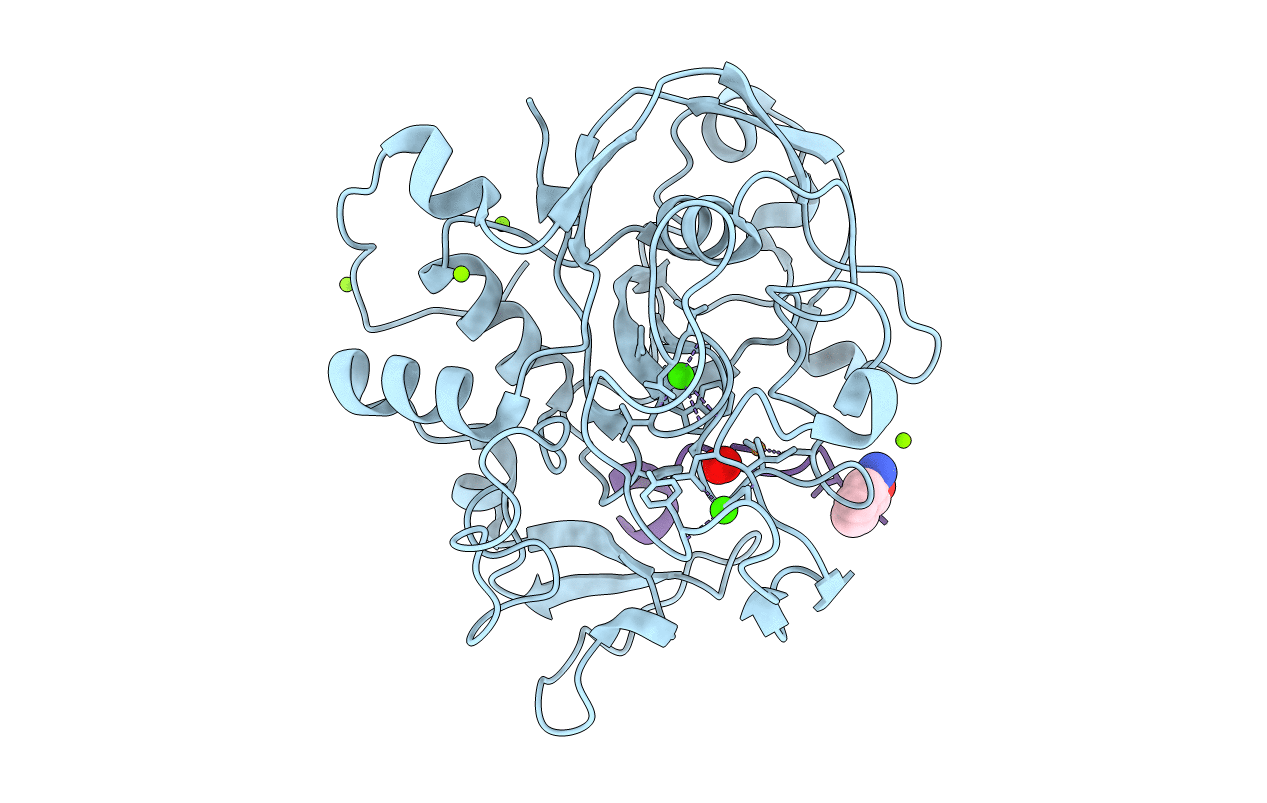
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1A Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, OXY, SOA |
 |
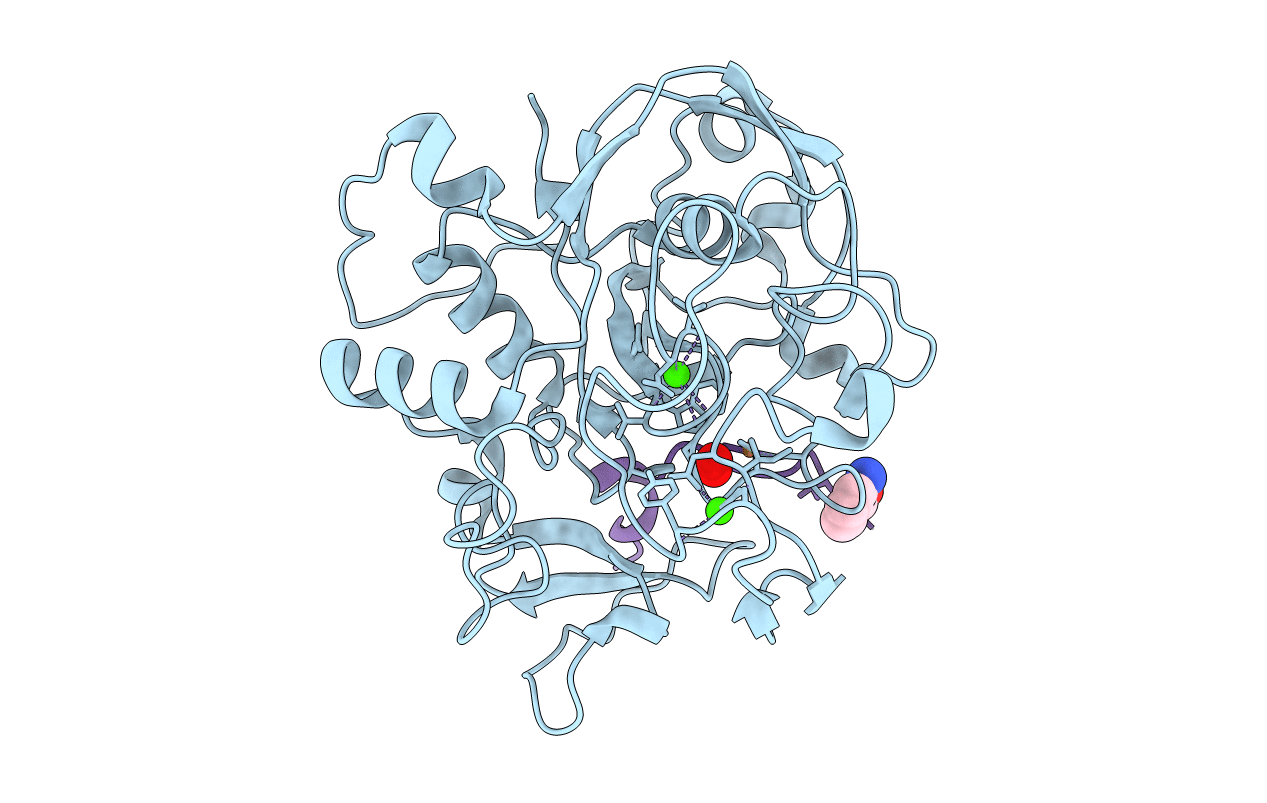
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1B Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, OXY, CL, MG, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1C Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, CL, OXY, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1D Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, CL, OXY, SOA |
 |
Native Crystal Structure Of Ergothioneine Degrading Enzyme Ergothionase From Treponema Denticola
Organism: Treponema denticola sp33
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-07-17 Classification: LYASE |
 |
Crystal Structure Of Ergothioneine Degrading Enzyme Ergothionase From Treponema Denticola In Complex With Desmethyl-Ergothioneine Sulfonic Acid
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-07-17 Classification: LYASE Ligands: KZ5 |
 |
Structure Of Formylglycine-Generating Enzyme At 1.04 A In Complex With Copper And Substrate Reveals An Acidic Pocket For Binding And Acti-Vation Of Molecular Oxygen.
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: CU1, CA, CL |
 |
Native Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, FMT, EOH, GOL |
 |
Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola In Persulfide Form.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, EDO, PEG, GOL |
 |
Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola In Complex With Natural Substrate Trimethyl Histidine.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: AVJ, NA, CL, SO4 |

