Search Count: 68
 |
Crystal Structure Of The Dimeric Transaminase Doed From C. Salexigens Dsm 3043.
Organism: Chromohalobacter israelensis dsm 3043
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: GOL, PLP |
 |
Biochemical And Structural Characterisation Of An Alkaline Family Gh5 Cellulase From A Shipworm Symbiont
Organism: Teredinibacter waterburyi
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: MG, TRS, EDO, K |
 |
The First Crystal Structure Of The Daba Aminotransferase Ectb In The Ectoine Biosynthesis Pathway Of The Model Organism Chromohalobacter Salexigens Dsm 3034
Organism: Chromohalobacter salexigens (strain dsm 3043 / atcc baa-138 / ncimb 13768)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-03-11 Classification: TRANSFERASE Ligands: PLP |
 |
Characterization Of An Intertidal Zone Metagenome Oligoribonuclease And The Role Of The Intermolecular Disulfide Bond For Homodimer Formation And Nuclease Activity.
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: MN |
 |
The Lipy/F-Motif In An Intracellular Subtilisin Protease Is Involved In Inhibition
Organism: Planococcus plakortidis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: PGE, NA, ACT |
 |
Discovery And Characterization Of A Thermostable Gh6 Endoglucanase From A Compost Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-06-06 Classification: HYDROLASE Ligands: EDO, SO4 |
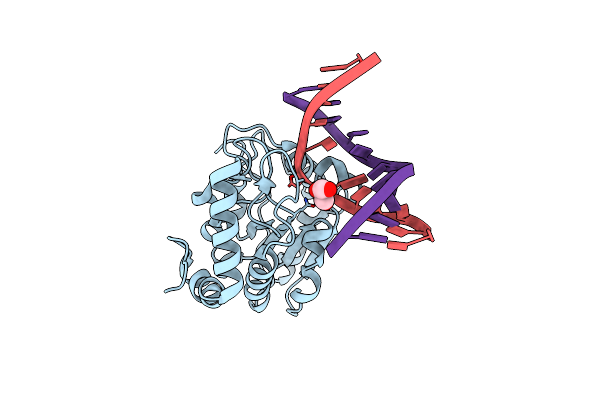 |
Crystal Structure Determination Of Uracil-Dna N-Glycosylase (Ung) From Deinococcus Radiodurans In Complex With Dna - New Insights Into The Role Of The Leucine-Loop For Damage Recognition And Repair
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-08-12 Classification: HYDROLASE/DNA Ligands: CL, GOL |
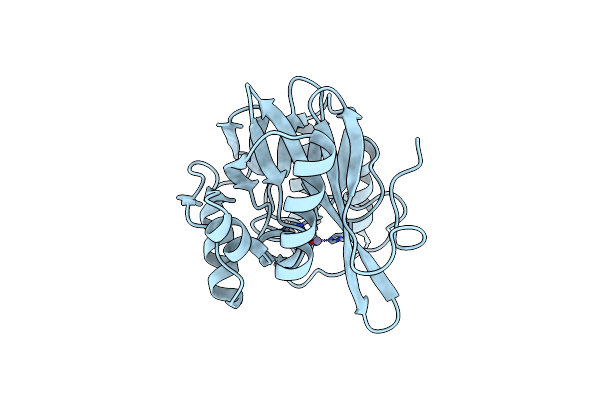 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
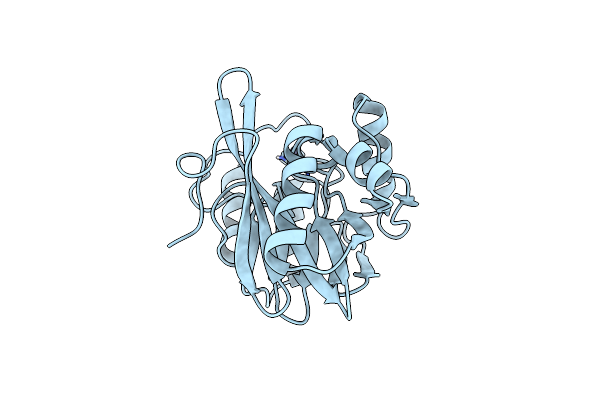 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
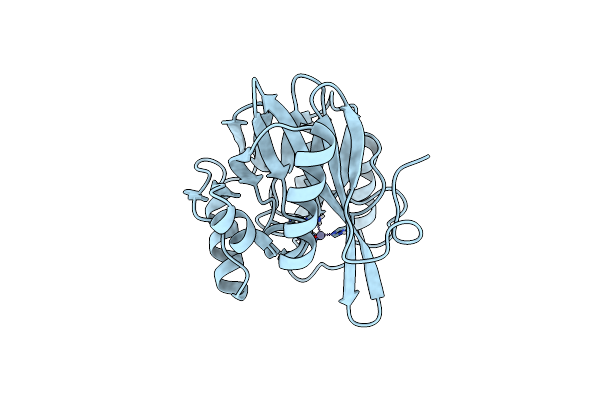 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Ctidh Bound To Nadp. The Complex Structures Of Isocitrate Dehydrogenase From Clostridium Thermocellum And Desulfotalea Psychrophila, Support A New Active Site Locking Mechanism
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-11 Classification: OXIDOREDUCTASE Ligands: ICT, MG, NAP |
 |
Dpidh-Nadp. The Complex Structures Of Isocitrate Dehydrogenase From Clostridium Thermocellum And Desulfotalea Psychrophila, Support A New Active Site Locking Mechanism
Organism: Desulfotalea psychrophila
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2012-07-11 Classification: OXIDOREDUCTASE Ligands: NAP, ICT, MG |
 |
Open Ctidh. The Complex Structures Of Isocitrate Dehydrogenase From Clostridium Thermocellum And Desulfotalea Psychrophila, Support A New Active Site Locking Mechanism
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2012-04-25 Classification: OXIDOREDUCTASE |
 |
Native Vim-7. Structural And Computational Investigations Of Vim-7: Insights Into The Substrate Specificity Of Vim Metallo-Beta- Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: ZN, MG, UNX |
 |
Vim-7 With Oxidised. Structural And Computational Investigations Of Vim-7: Insights Into The Substrate Specificity Of Vim Metallo-Beta- Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: ZN, MG, UNX |
 |
Vim-7 With Oxidised. Structural And Computational Investigations Of Vim-7: Insights Into The Substrate Specificity Of Vim Metallo-Beta- Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of An Unusual 3-Methyladenine Dna Glycosylase Ii (Alka) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: CL, GOL, MES |
 |
Structure Of An Unusual 3-Methyladenine Dna Glycosylase Ii (Alka) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: CL, GOL, NI |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-05-12 Classification: HYDROLASE/INHIBITOR Ligands: ILE, TYR, ZN, CA, PEG |

