Search Count: 15
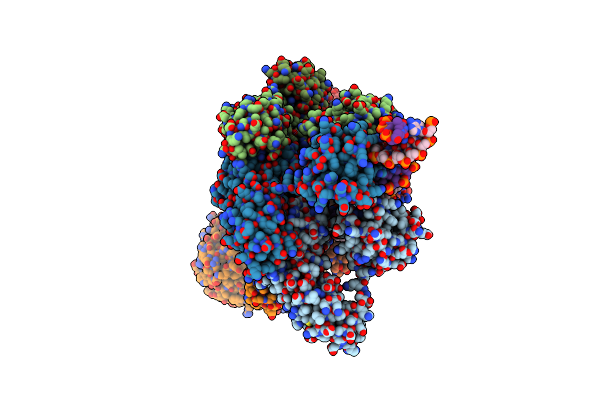 |
Cryo-Em Structure Of The Bacterial Replication Origin Opening Basal Unwinding System
Organism: Bacillus subtilis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: REPLICATION Ligands: MG, ATP |
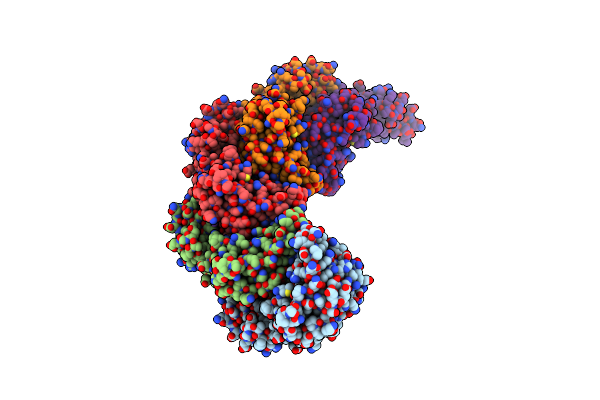 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: ANP, K, MG |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-09-27 Classification: FLUORESCENT PROTEIN |
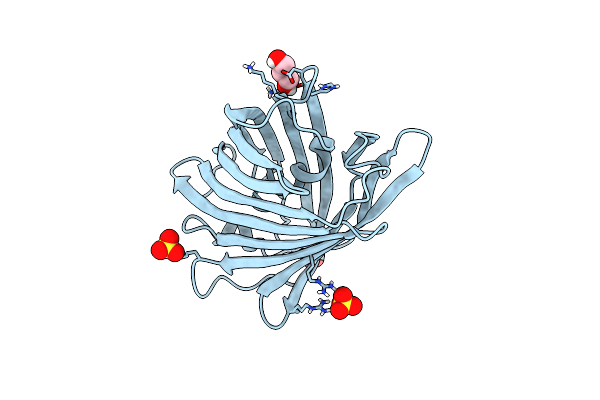 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
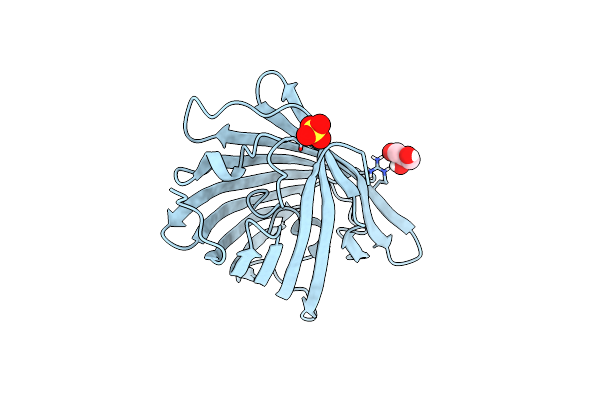 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2022-11-09 Classification: FLUORESCENT PROTEIN Ligands: SO4 |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-08-07 Classification: CELL CYCLE Ligands: BR |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-03-02 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2011-03-02 Classification: TRANSCRIPTION Ligands: PO4 |

