Search Count: 14
 |
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-12-26 Classification: TRANSFERASE Ligands: ANP, M7B, K, CL, PO4, MG, GMZ |
 |
Crystal Structure Of Burkholderia Cenocepacia Hlda In Complex With An Atp-Competitive Inhibitor
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2012-12-26 Classification: Transferase/Transferase Inhibitor Ligands: IHA, K |
 |
Crystal Structure Of Burkholderia Cenocepacia Hlda In Complex With An Atp-Competitive Inhibitor
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-12-26 Classification: Transferase/Transferase Inhibitor Ligands: IHA, M7B, K, CL, PO4 |
 |
Crystal Structure Of Burkholderia Cenocepacia Hlda In Complex With An Atp-Competitive Inhibitor
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2012-12-26 Classification: Transferase/Transferase Inhibitor Ligands: IHC, K |
 |
Crystal Structure Of The Ternary Complex Between S100A10, An Annexin A2 N-Terminal Peptide And An Ahnak Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-10-24 Classification: EXOCYTOSIS/PROTEIN BINDING |
 |
Crystal Structure Of The Second Tetrahedral Intermediates Of Sgpb At Ph 4.2
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2007-12-11 Classification: HYDROLASE Ligands: CL, SO4, EDO, GOL |
 |
Crystal Structure Of The Second Tetrahedral Intermediates Of Sgpb At Ph 7.3
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2007-12-11 Classification: HYDROLASE Ligands: SO4, CL, TYR, LEU, EDO, EPE, GOL, ACY |
 |
Organism: Meleagris gallopavo
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2007-02-13 Classification: HYDROLASE INHIBITOR Ligands: CL |
 |
Crystal Structure Of The P14'-Ala32 Variant Of The N-Terminally Truncated Omtky3-Del(1-5)
Organism: Meleagris gallopavo
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2007-02-13 Classification: HYDROLASE INHIBITOR |
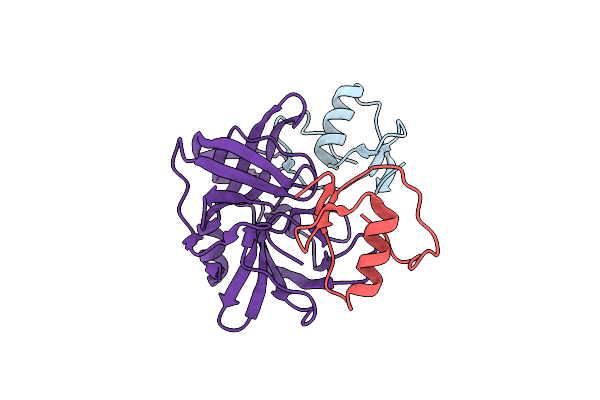 |
Organism: Meleagris gallopavo, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-02-13 Classification: HYDROLASE/HYDROLASE INHIBITOR |
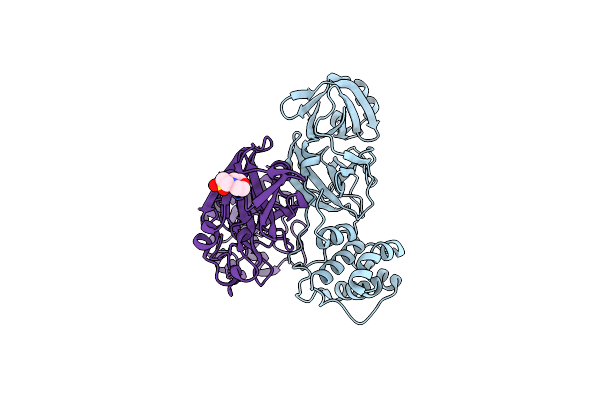 |
Crystal Structure Of Sars Coronavirus Main Peptidase At Ph 6.0 In The Space Group P21
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: MES |
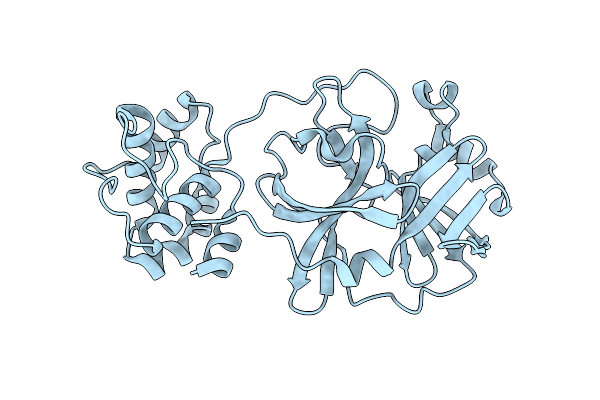 |
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE |
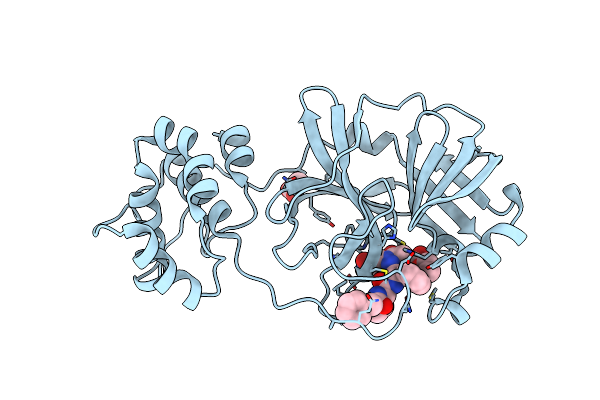 |
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) Inhibited By An Aza-Peptide Epoxide In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: AZP, ACY |
 |

