Search Count: 35
 |
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Organism: Klebsiella pneumoniae, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The G15C-R66C And T83C-T83C Diabody Complex (Cits-Diabody #7-Tlr3)
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The Klebsiella Pneumoniae Cits (Citrate-Bound Occluded State)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: CIT, PLM |
 |
Cryo-Em Structure Of The Human Glucose Transporter, Glut7 In Outward-Facing Open Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: TRANSPORT PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: GSH, CD |
 |
Cryo-Em Structure Of The Human Abcb6 In Complex With Cd(Ii):Phytochelatin 2
Organism: Homo sapiens, Tracheophyta
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: CD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSPORT PROTEIN Ligands: PEV, AOV, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSPORT PROTEIN Ligands: ADP, VO4, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: Y01, HT9 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: GSH, Y01, HEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: CHD, A3P |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-09-11 Classification: PROTEIN BINDING |
 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: GOL, 9KF |
 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: GOL, 9KL |
 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-13 Classification: HYDROLASE |
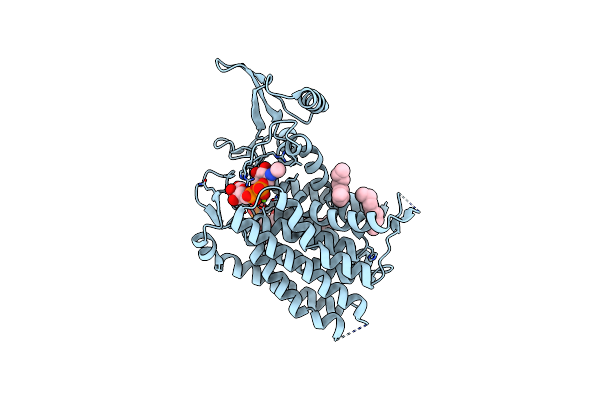 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) (V264G Mutant) In Complex With Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-07-25 Classification: TRANSFERASE Ligands: MG, UD1, UNL, P6L |
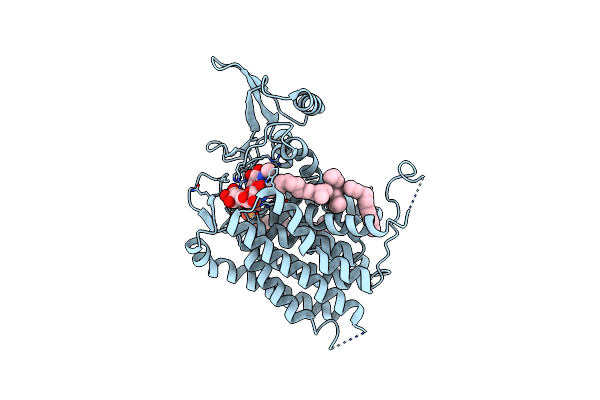 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) (V264G Mutant) In Complex With Tunicamycin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: P6L, 9LH, UNL |
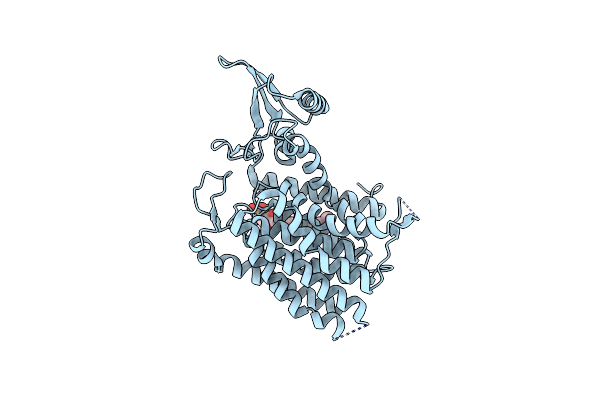 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: P6L |
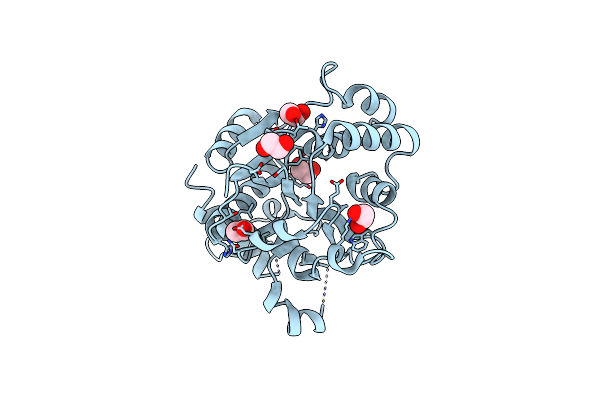 |
Crystal Structure Of Human Glycogenin-1 (Gyg1) Tyr195Piphe Mutant, Apo Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: EDO |

