Search Count: 1,886
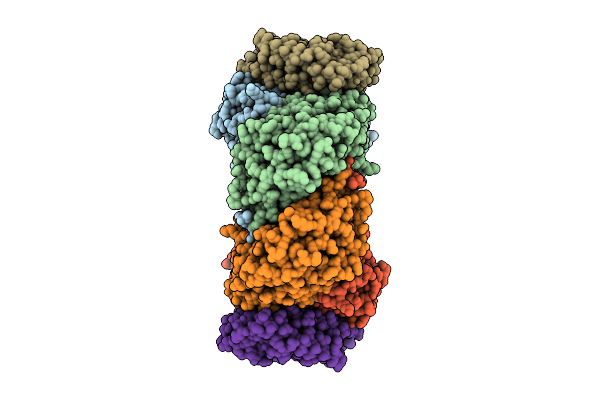 |
Cryo-Em Structure Of Soluble Methane Monooxygenase Hydroxylase From Methylosinus Sporium 5
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: FE |
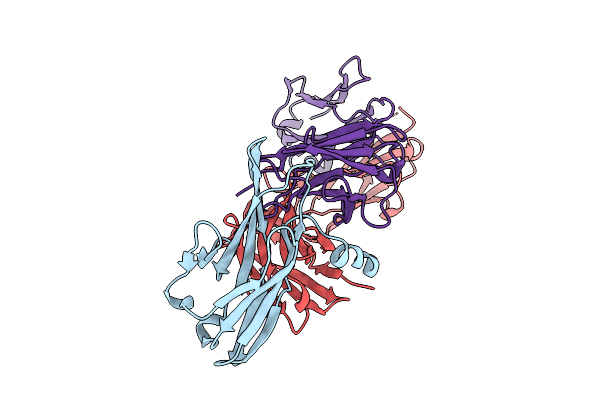 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA |
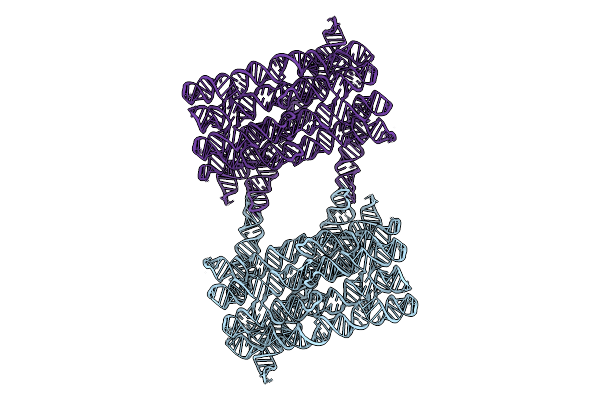 |
1-Methyl-Pseudouridine Twist Corrected Rna Origami 6-Helix Bundle Type-1 Dimer
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA |
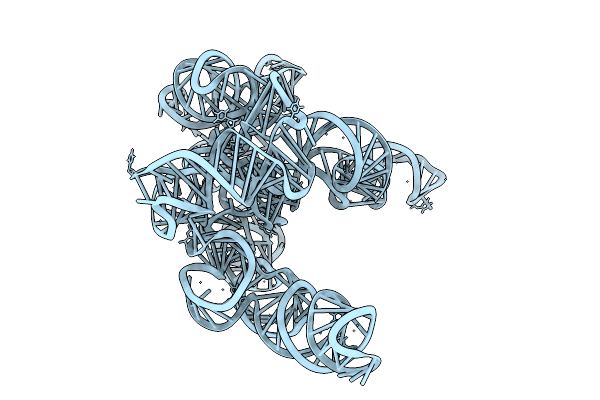 |
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
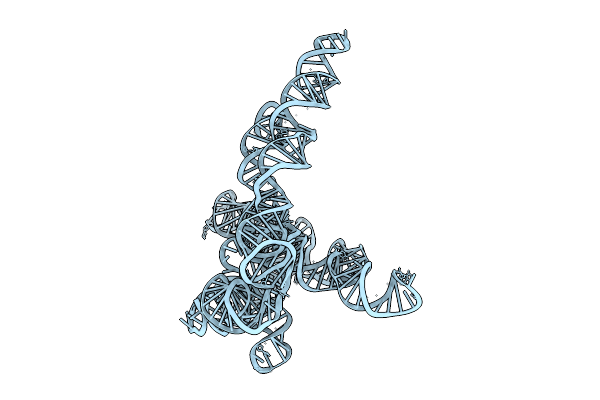 |
1-Methyl-Pseudouridine L-21 Scai Tetrahymena Ribozyme - Extended Conformation
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
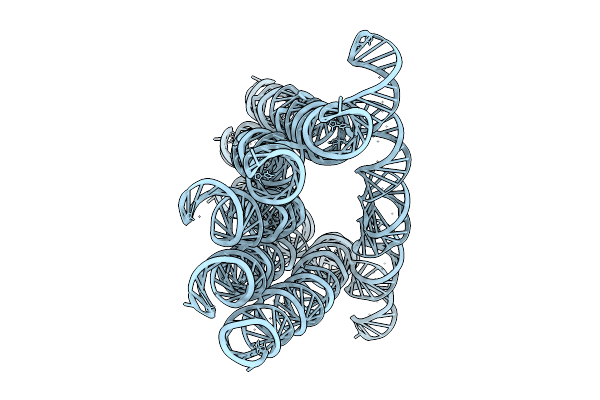 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
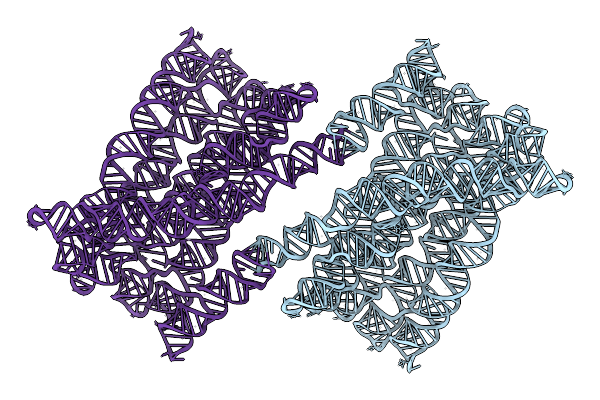 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
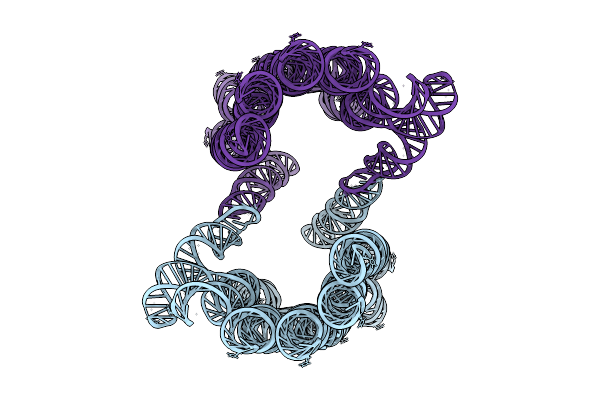 |
1-Methyl-Pseudouridine Twist Corrected Rna Origami 6-Helix Bundle Type-2 Dimer
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
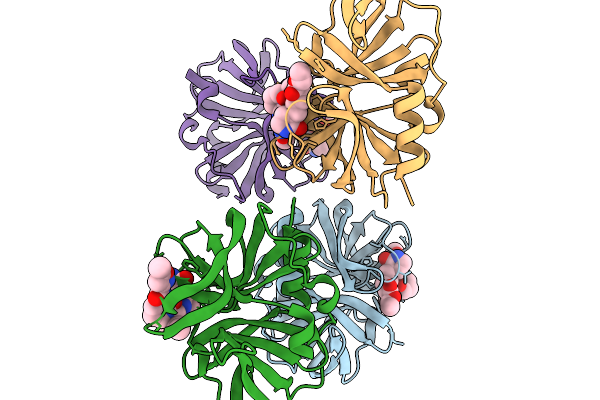 |
Organism: Rhinovirus b14
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: XNV |
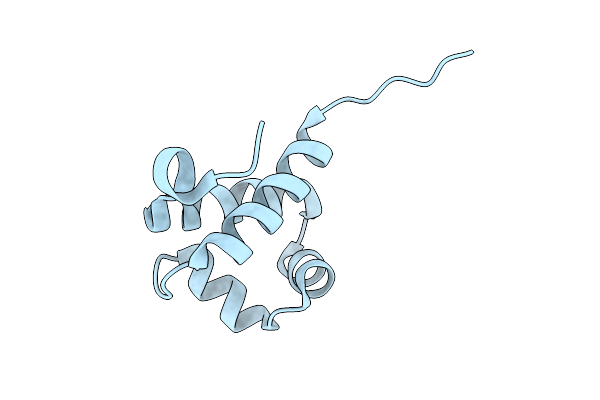 |
Organism: Halomonas caseinilytica
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN |
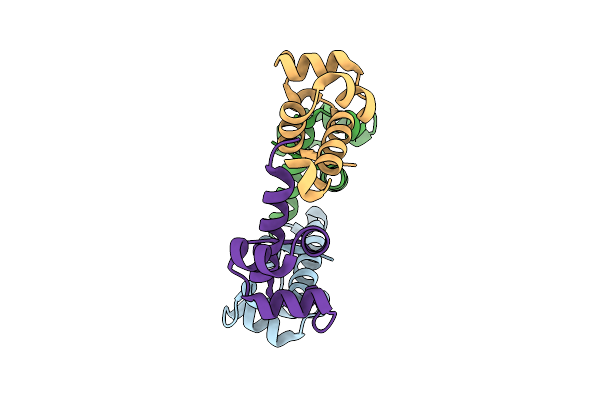 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN |
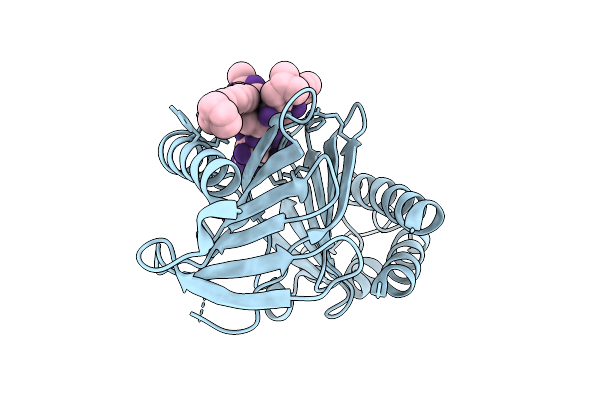 |
Organism: Mus musculus, Synthetic constrcut
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE |
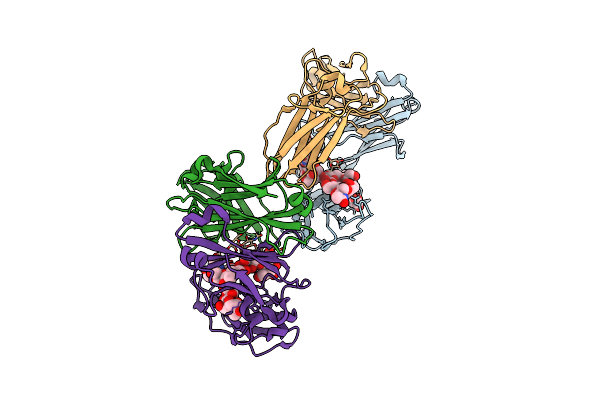 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
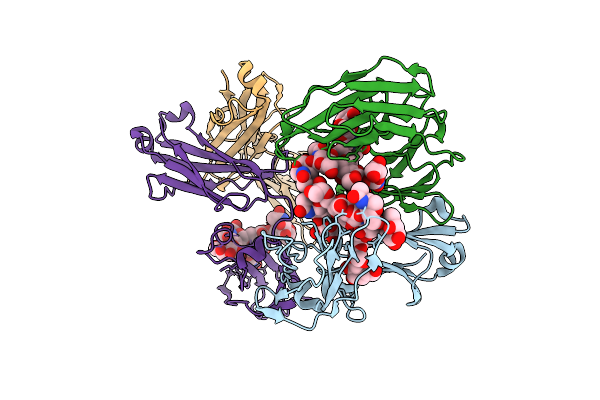 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: FUL |
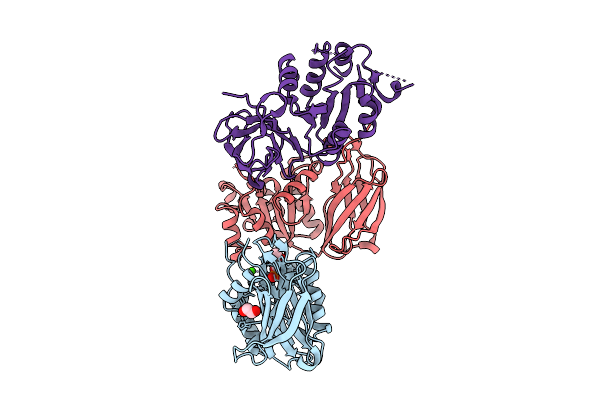 |
Crystal Structure Of Nir2 C-Terminal Domain In Complex With Phosphatidic Acid
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: LIPID BINDING PROTEIN Ligands: 44E, DW3, CA |
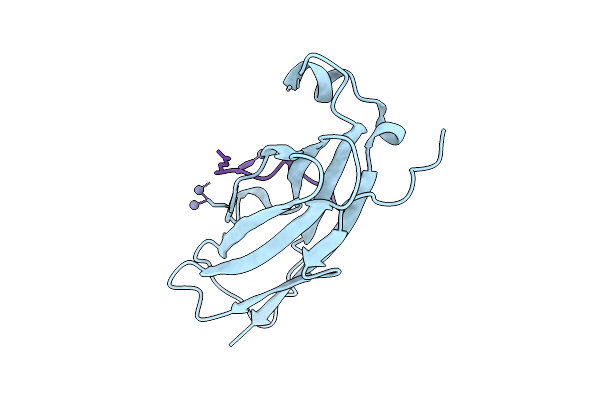 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: LIPID BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: LIPID BINDING PROTEIN Ligands: PO4 |

