Search Count: 97
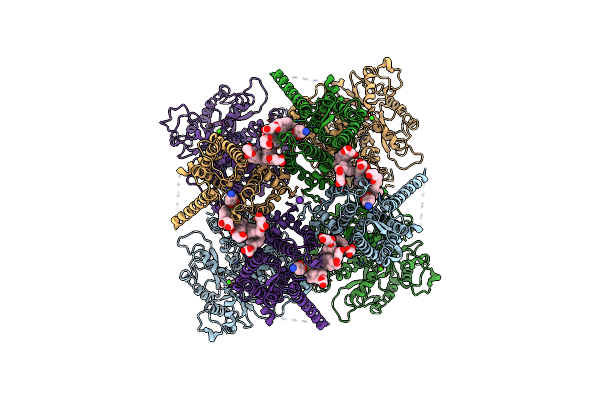 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: MG, CA, K, A1L4E, Y01 |
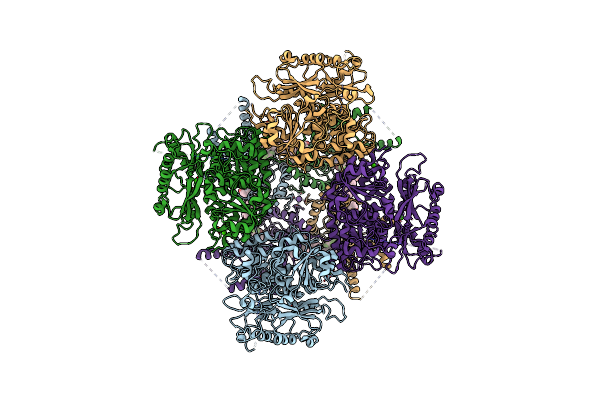 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: A1L4F, MG, CA, K, Y01 |
 |
Crystal Structure Of Polyketide Synthase (Pks) Thioreductase Domain From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, GOL, BTB |
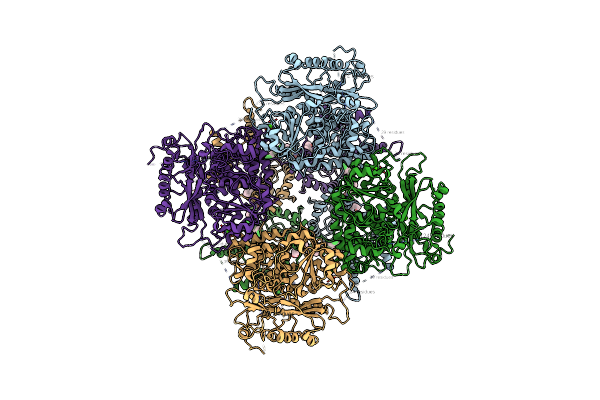 |
Activation Mechanism And Novel Binding Site Of The Bkca Channel Activator Ctibd
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: A1L04, Y01, MG, CA |
 |
Organism: Bacillus amyloliquefaciens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2024-06-05 Classification: PROTEIN BINDING |
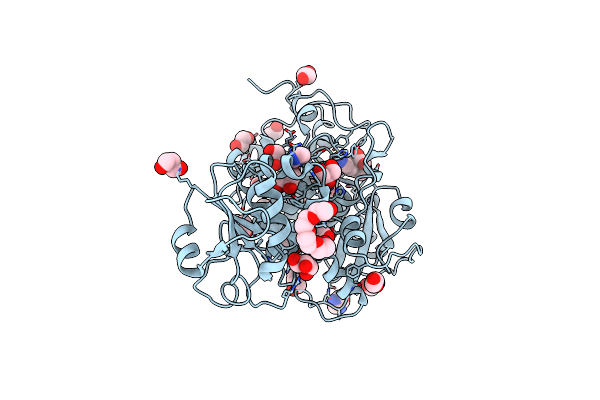 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: AVJ, MG, CL, GOL, EDO, PEG, PG4, PDO, IMD, NI |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, CL, PGE, GOL, IMD, NA, EDO |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, CL, PEG, MPO, EDO, PGE |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, PEG, EDO, CL, BR |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, LW8, CL, EDO |
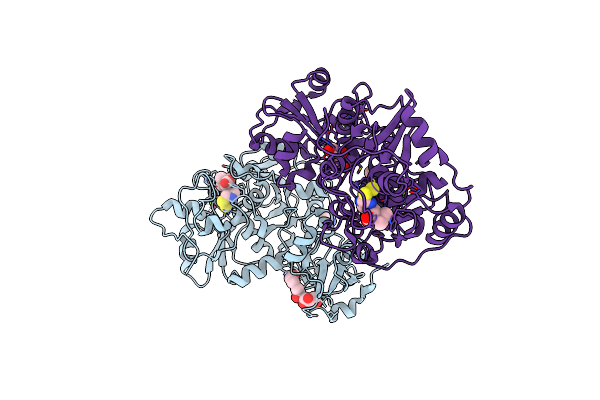 |
The Structure Of Eanb/Y353A Complex With Ergothioneine Covalent Linked With Persulfide Cys412
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: DV6, MG, CL, GOL, IMD, ZN, BR, PEG, EDO |
 |
Crystal Structure Of Genb1 From Micromonospora Echinospora In Complex With Plp (Internal Aldimine)
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: PLP, MG |
 |
Crystal Structure Of Genb1 From Micromonospora Echinospora In Complex With Ji-20A And Plp (External Aldimine)
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: PLP, JIA |
 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: PLP, XXX |
 |
Crystal Structure Of Carbon Sulfoxide Lyase, Egt2 Y134F With Sulfenic Acid Intermediate
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-03-07 Classification: LYASE Ligands: FMT, 8QJ |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2018-03-07 Classification: LYASE Ligands: FMT, 8VV |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-02-21 Classification: LYASE Ligands: FMT |
 |
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-06-14 Classification: PROTEIN TRANSPORT |
 |
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2017-06-14 Classification: PROTEIN TRANSPORT Ligands: ZN |
 |
Structure Of The Cytosolic Domain Of Dotm Derived From Legionella Pneumophila
Organism: Legionella pneumophila (strain lens)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-14 Classification: PROTEIN TRANSPORT |

