Search Count: 47
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YK, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YV, CL, MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-18 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-09-18 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In Apo Form
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD |
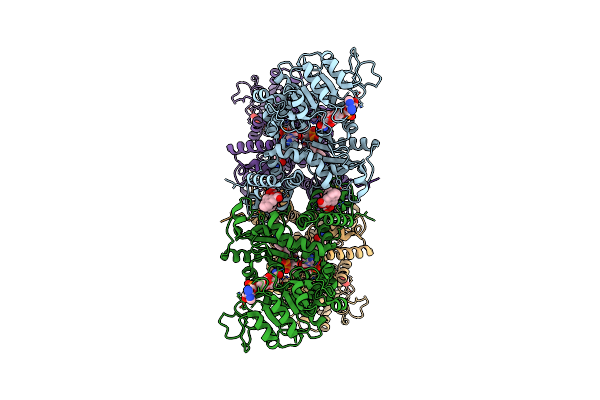 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Ea
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, JBR |
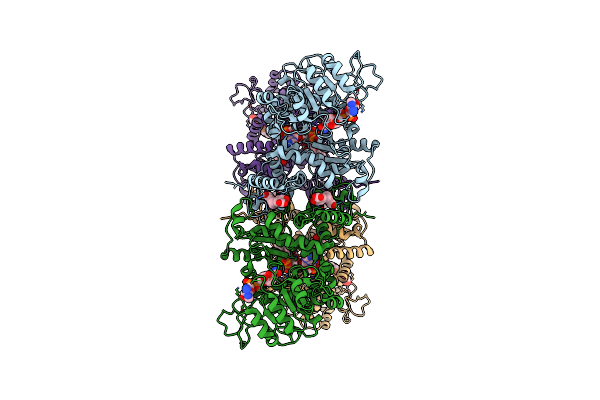 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Mdsa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, D5S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: 5YS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN Ligands: QY1, CA, BOG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN Ligands: QY7, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-12-02 Classification: ISOMERASE, MEMBRANE PROTEIN Ligands: QY1, BOG, PG4 |
 |
Crystal Structure Of Human Pim-1 Kinase In Complex With A Quinazolinone-Pyrrolodihydropyrrolone Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: JYG, GOL |
 |
Organism: Opisthorchis viverrini
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-10-24 Classification: ISOMERASE Ligands: MG |
 |
Crystal Structure Of Triosephosphate Isomerase Sad Deletion Mutant From Opisthorchis Viverrini
Organism: Opisthorchis viverrini
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-10-24 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase Sadsubaaa Mutant From Opisthorchis Viverrini
Organism: Opisthorchis viverrini
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-10-24 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase Sad Deletion And N115A Mutant From Opisthorchis Viverrini
Organism: Opisthorchis viverrini
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-10-24 Classification: ISOMERASE |
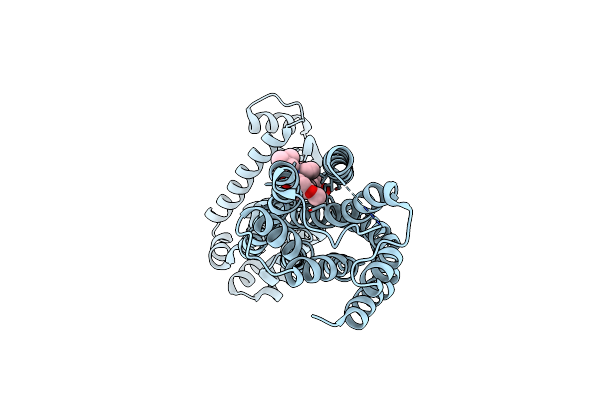 |
The Extra-Helical Binding Site Of Gpr40 And The Structural Basis For Allosteric Agonism And Incretin Stimulation
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2018-05-02 Classification: Fatty acid binding protein/Hydrolase Ligands: 6XQ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: 8UM, AMZ, MG |
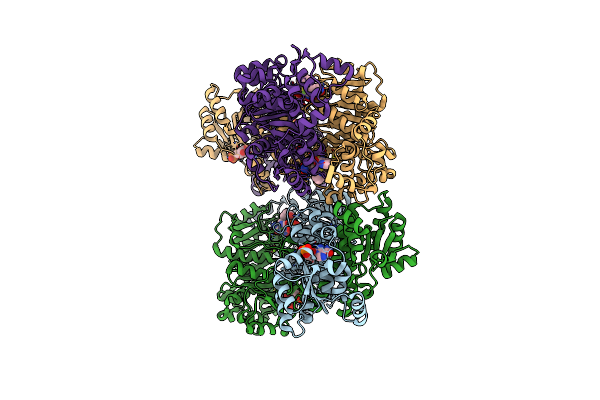 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: AMZ, MG, 8US |

