Search Count: 16
 |
Organism: Gelidibacter salicanalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: FAD |
 |
Organism: Gelidibacter salicanalis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: NDP, FAD |
 |
Organism: Caballeronia sordidicola
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2023-12-06 Classification: HYDROLASE |
 |
Organism: Bosea sp. pamc 26642
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-11-22 Classification: HYDROLASE |
 |
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-17 Classification: HYDROLASE |
 |
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-05-17 Classification: HYDROLASE Ligands: ZYC |
 |
Organism: Variovorax sp.
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-10-12 Classification: HYDROLASE |
 |
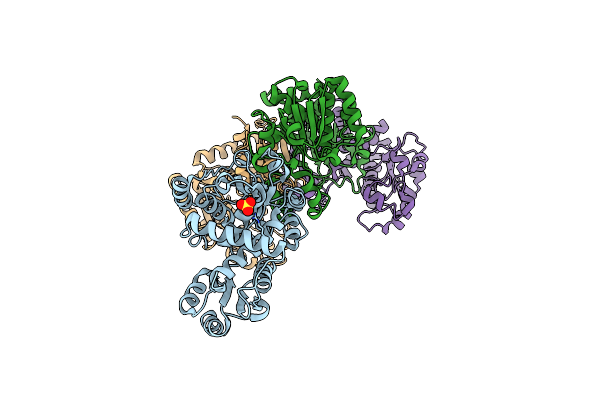
Anabolic Ornithine Carbamoyltransferases (Otcs) From Psychrobacter Sp. Pamc 21119
Organism: Psychrobacter sp. pamc 21119
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-31 Classification: TRANSFERASE |
 |
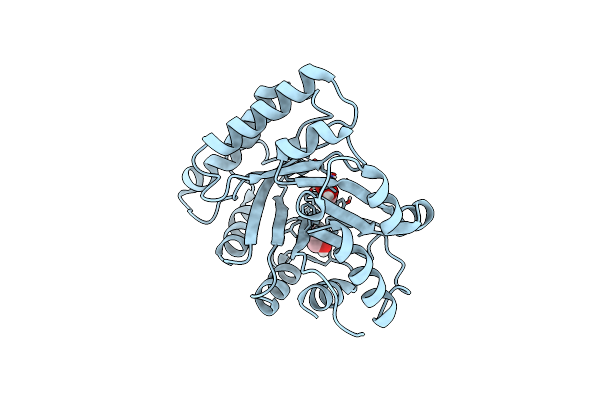
Catabolic Ornithine Carbamoyltransferases (Otcs) From Psychrobacter Sp. Pamc 21119
Organism: Psychrobacter sp. pamc 21119
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: SO4 |
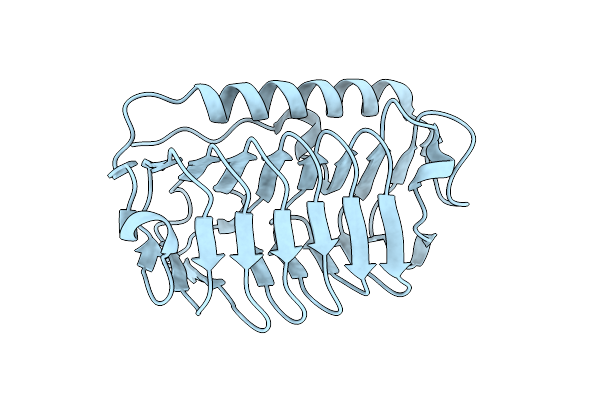 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-02 Classification: ANTIFREEZE PROTEIN Ligands: CL |
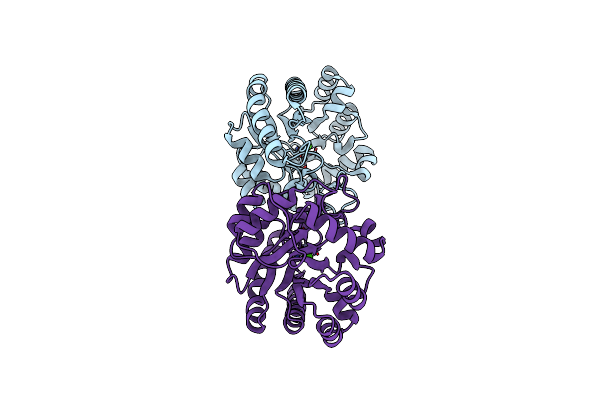 |
Crystal Structure Of A Novel Putative Sugar Isomerase From The Psychrophilic Bacterium Paenibacillus Sp. R4
Organism: Paenibacillus sp. fsl h7-0331
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2021-12-29 Classification: ISOMERASE Ligands: CA, ZN |
 |
Crystal Structure Of Phosphoenolpyruvate Carboxylase From Arabidopsis Thaliana In Complex With Aspartate And Citrate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-02-08 Classification: LYASE Ligands: ASP, FLC |
 |
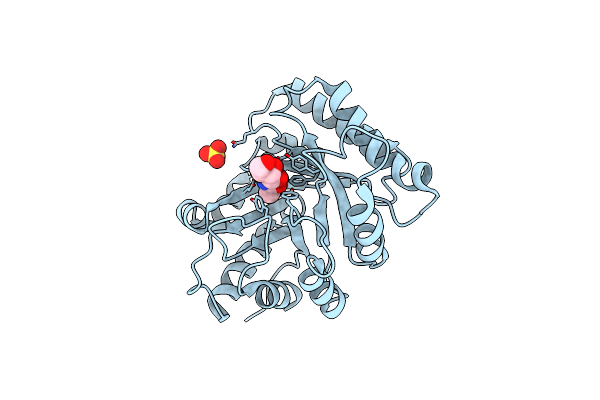
Organism: Aspergillus clavatus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: EDO |
 |
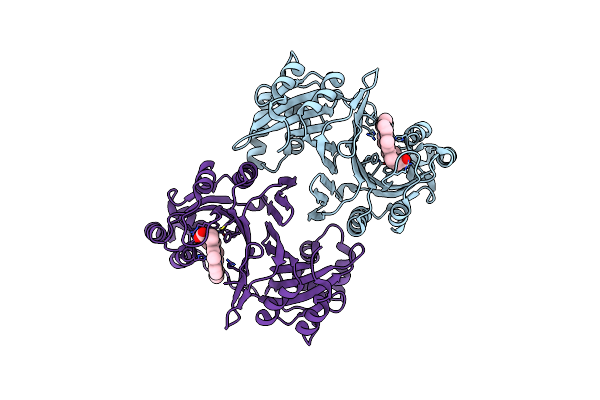
Organism: Aspergillus clavatus (strain atcc 1007 / cbs 513.65 / dsm 816 / nctc 3887 / nrrl 1)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: CL, SO4, NGA |
 |
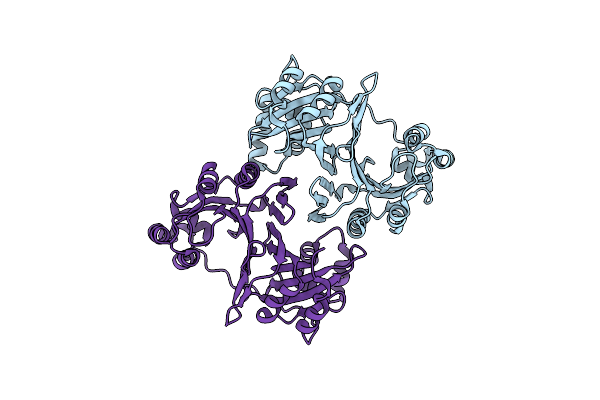
Crystal Structure Of Holo-Phus, A Heme-Binding Protein From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-03-05 Classification: METAL TRANSPORT Ligands: HEM |
 |
Crystal Structure Of Apo-Phus, A Heme-Binding Protein From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-03-05 Classification: METAL TRANSPORT |

