Search Count: 14
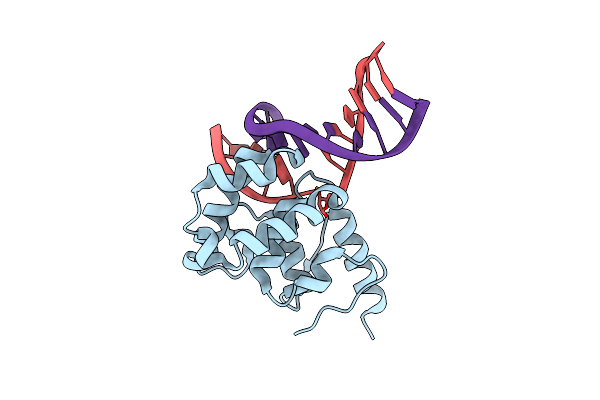 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2021-01-27 Classification: TRANSFERASE |
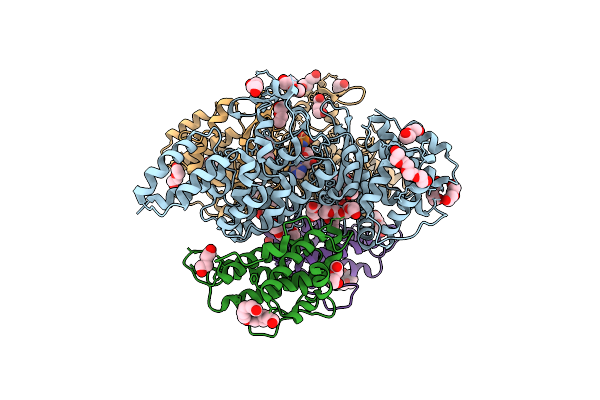 |
L. Pneumophila Effector Kinase Legk7 (Amp-Pnp Bound) In Complex With Human Mob1A
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: PG4, PEG, P6G, ZN, MN, ANP |
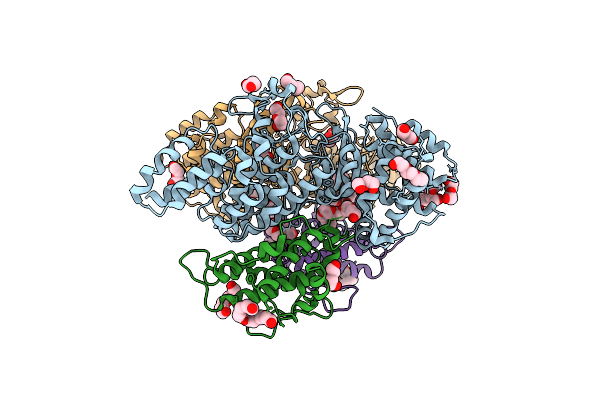 |
Organism: Legionella pneumophila subsp. pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: PEG, P6G, PG4, ZN |
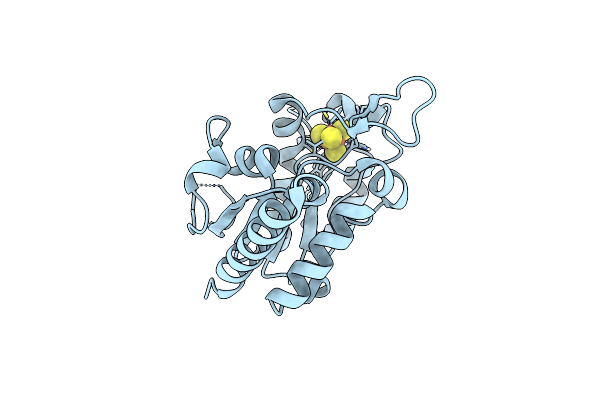 |
Crystal Structure At 1.3 Angstroms Resolution Of A Novel Udg, Udgx, From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, PO4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: DU, SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-05-25 Classification: TRANSFERASE Ligands: CAQ, SO4, NH4 |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2012-10-31 Classification: HYDROLASE |
 |
Complex Membrane Type-1 Matrix Metalloproteinase (Mt1-Mmp) With Tissue Inhibitor Of Metalloproteinase-1 (Timp-1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-06-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, ZN |
 |
The Crystal Structure Of An Acremonium Strictum Glucooligosaccharide Oxidase Reveals A Novel Flavinylation
Organism: Acremonium strictum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAG, ZN, FAD |
 |
Crystal Structure Of Glucooligosaccharide Oxidase From Acremonium Strictum: A Novel Flavinylation Of 6-S-Cysteinyl, 8Alpha-N1-Histidyl Fad
Organism: Acremonium strictum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAG, ZN, FAD, ABL |

