Search Count: 730
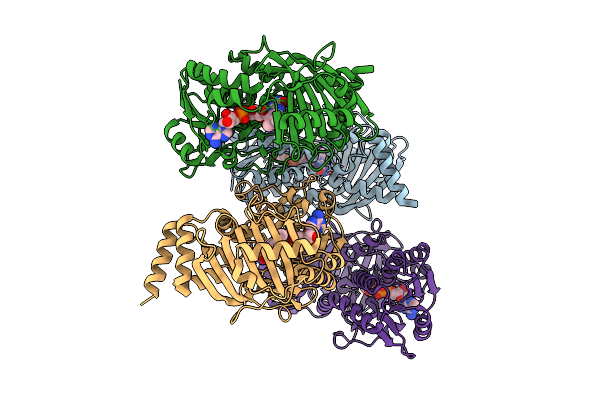 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: FAD |
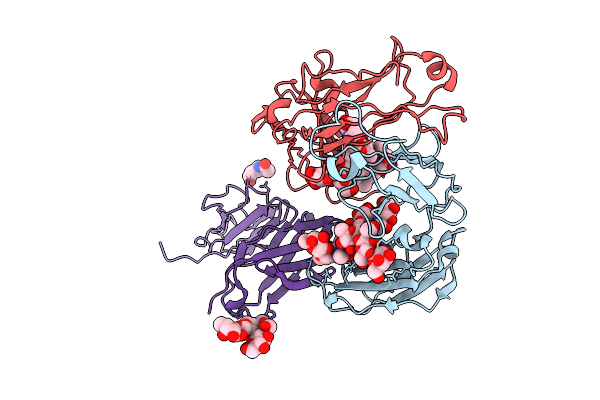 |
Crystal Structure Of Rhesus Macaque (Macaca Mulatta) Igg1 Fc Fragment- Fc-Gamma Receptor Iia Complex P131 Variant
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: NAG |
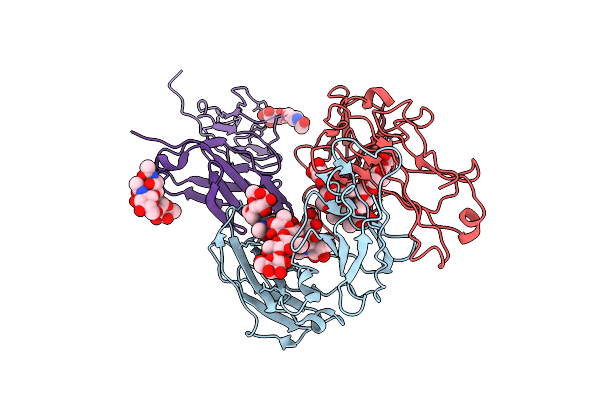 |
Crystal Structure Of Rhesus Macaque (Macaca Mulatta) Igg1 Fc Fragment- Fc-Gamma Receptor Iia Complex H131 Variant
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM |
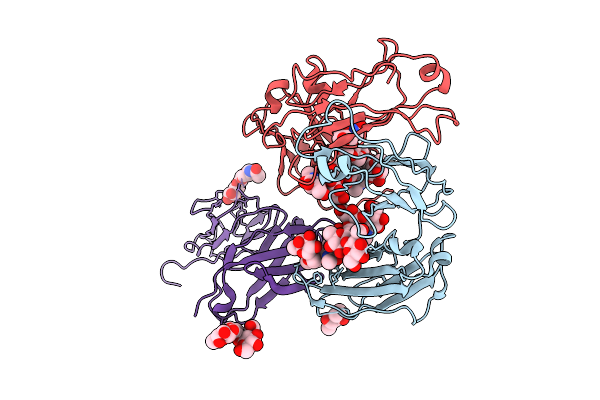 |
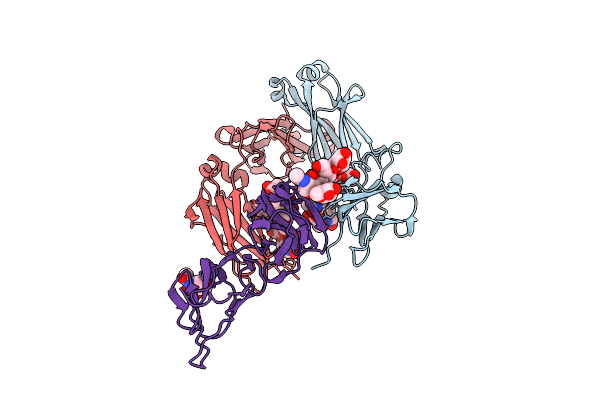
Crystal Structure Of Rhesus Macaque (Macaca Mulatta) Igg2 Fc Fragment- Fc-Gamma Receptor Iia Complex H131 Variant
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: MPD |
 |
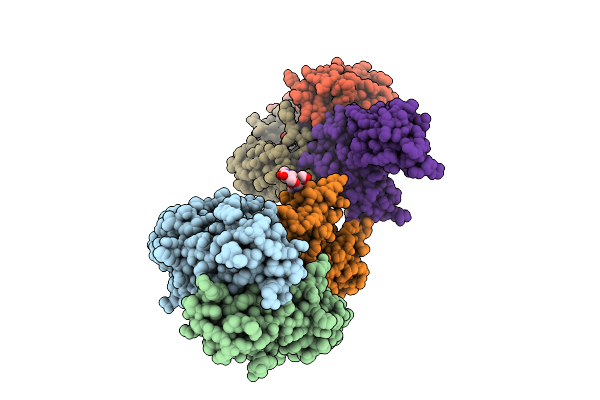
Crystal Structure Of Human Igg1 Fc Fragment-Fc-Gamma Receptor Iia Complex H131 Variant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
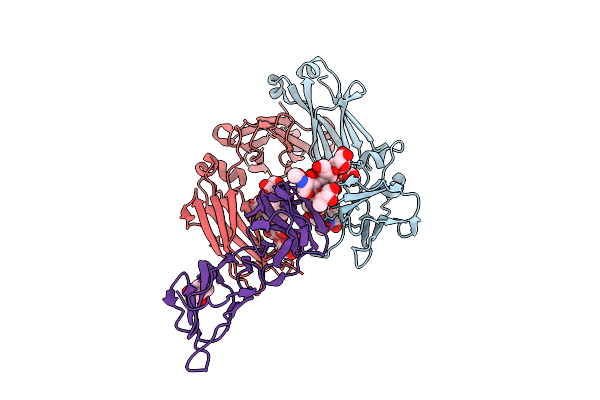
Crystal Structure Of Human Igg1 Fc Fragment-Fc-Gamma Receptor Iia Complex R131 Variant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: NAG, MPD |
 |
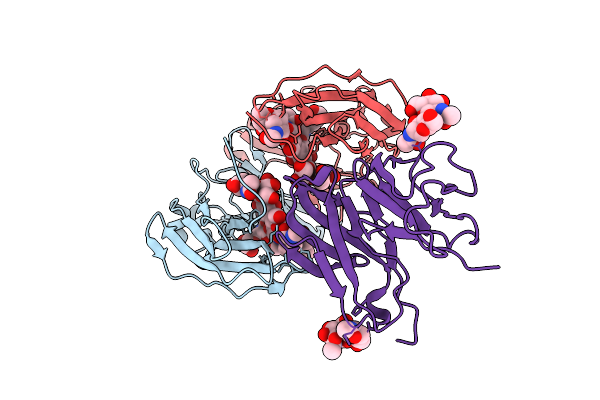
Crystal Structure Of Human Igg2 Fc Fragment-Fc-Gamma Receptor Iia Complex H131 Variant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: HOH |
 |
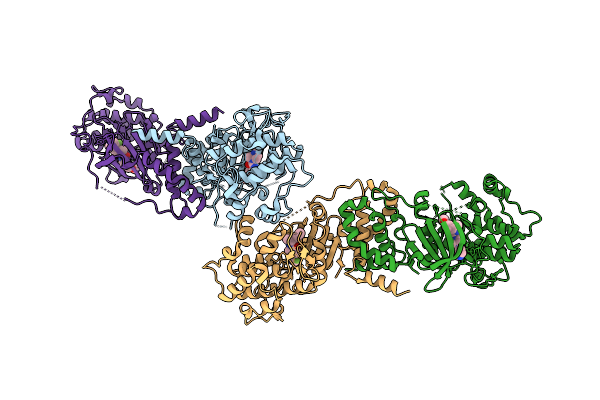
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YK, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YV, CL, MES |
 |
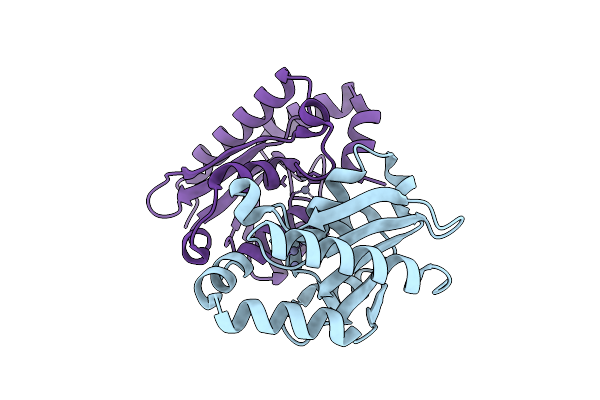
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YO |
 |
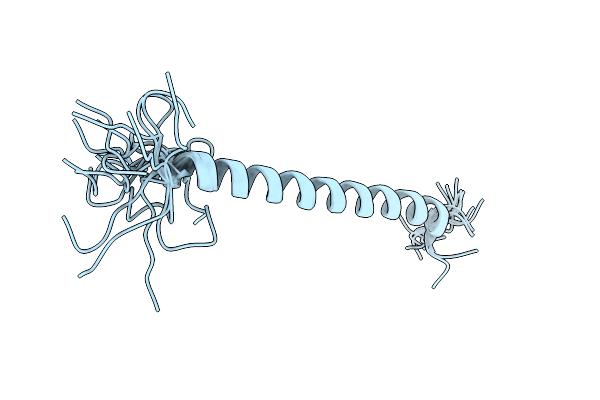
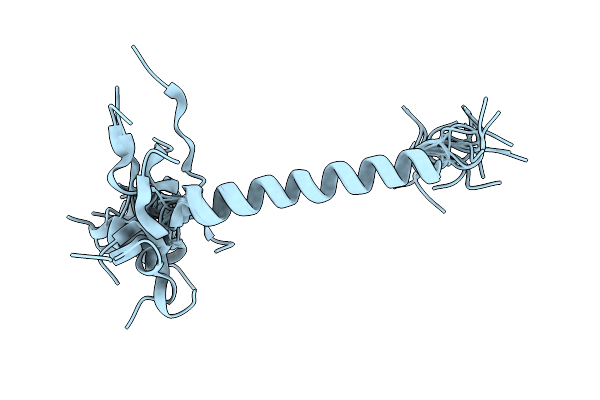
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
 |
 |
High-Resolution Crystal Structure Of Vibrio Cholerae Nfeob In The Gdp-Bound Form
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GDP, GOL |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GCP, GOL, MG |
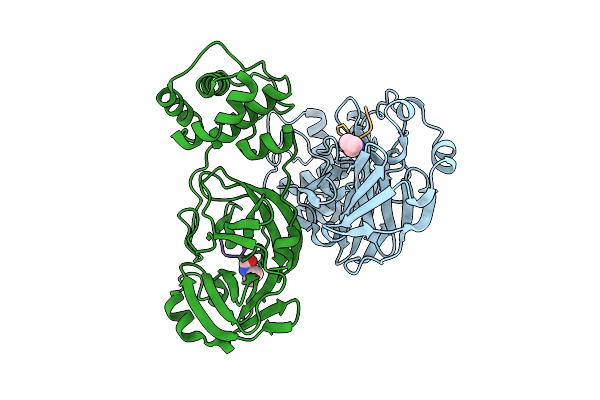 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
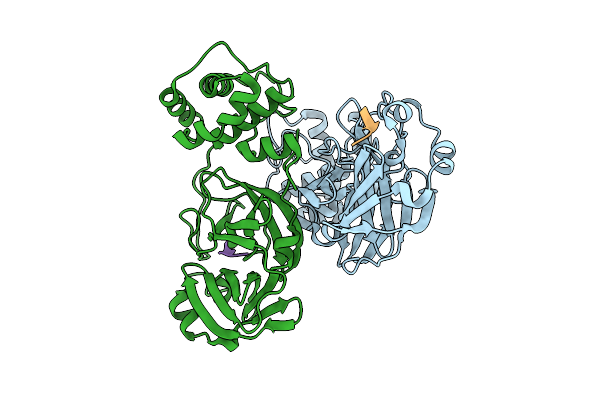 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
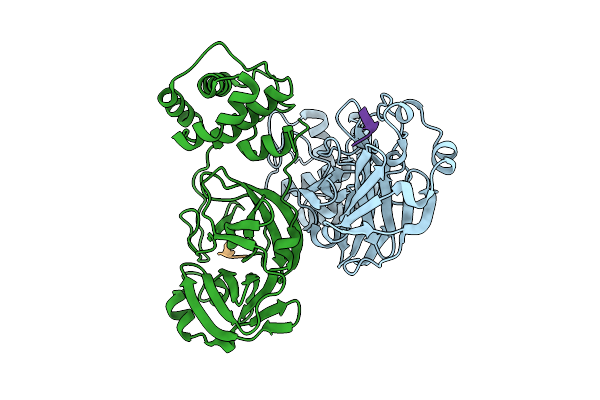 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
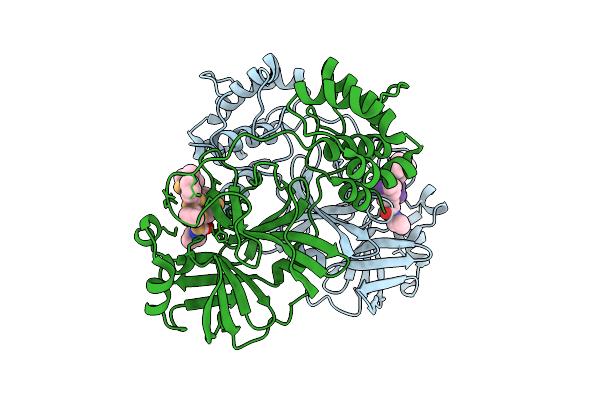 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |

