Search Count: 561
 |
Crystal Structure Of N-Terminal Domain Of Hypothetical Protein Rv1421 From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: P6G |
 |
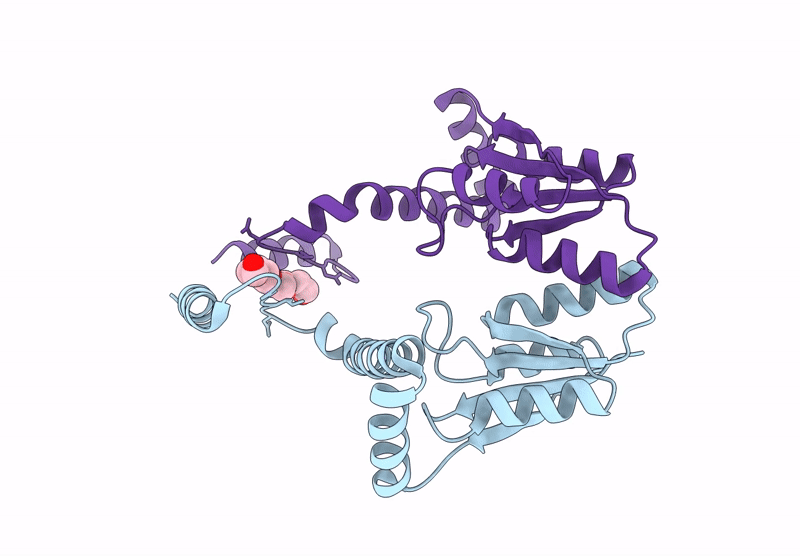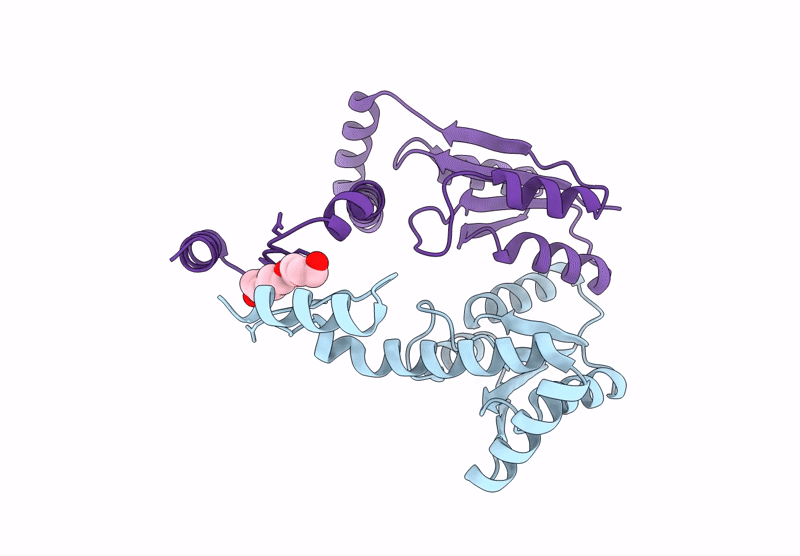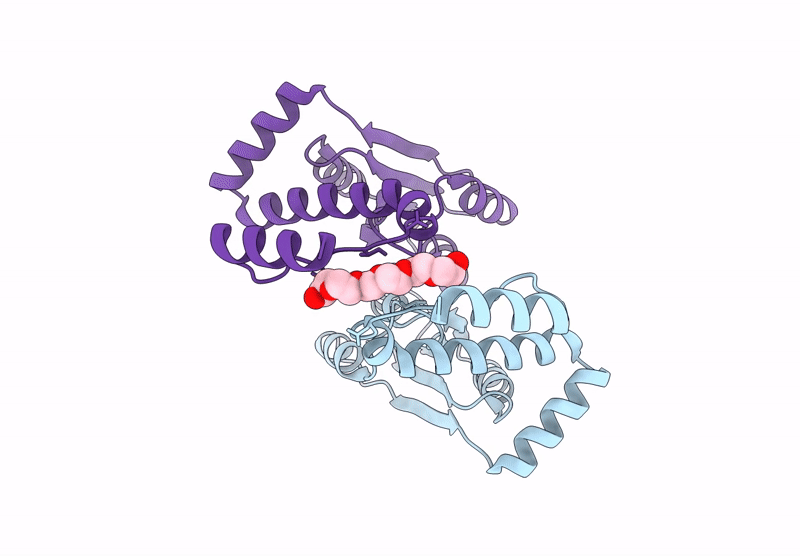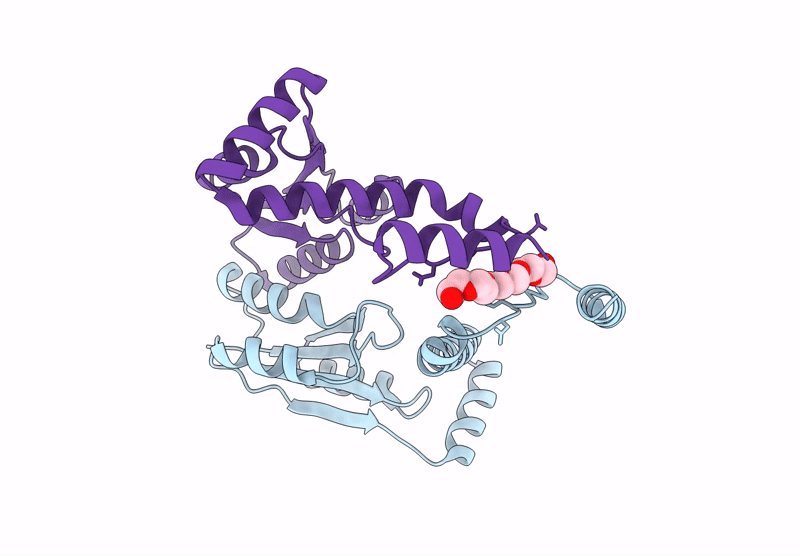
Crystal Structure Of The N-Terminal Kinase Domain From Saccharomyces Cerevisiae Vip1 In Complex With Adp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, EDO |
 |
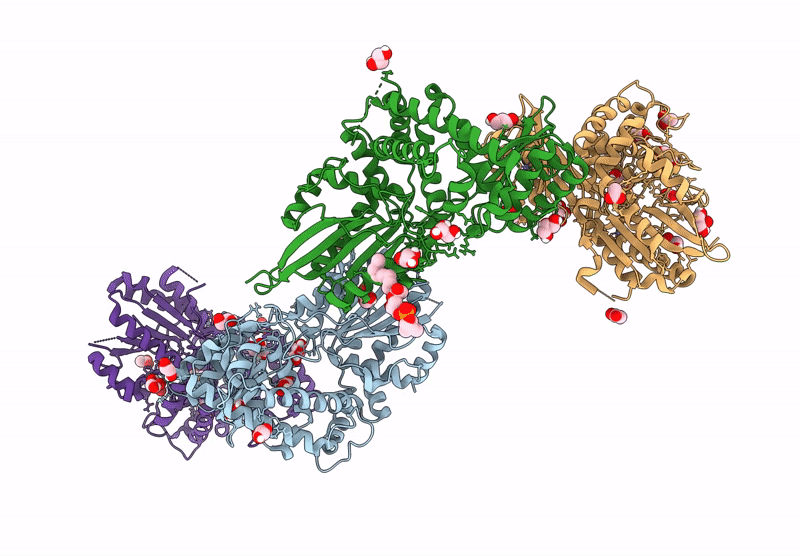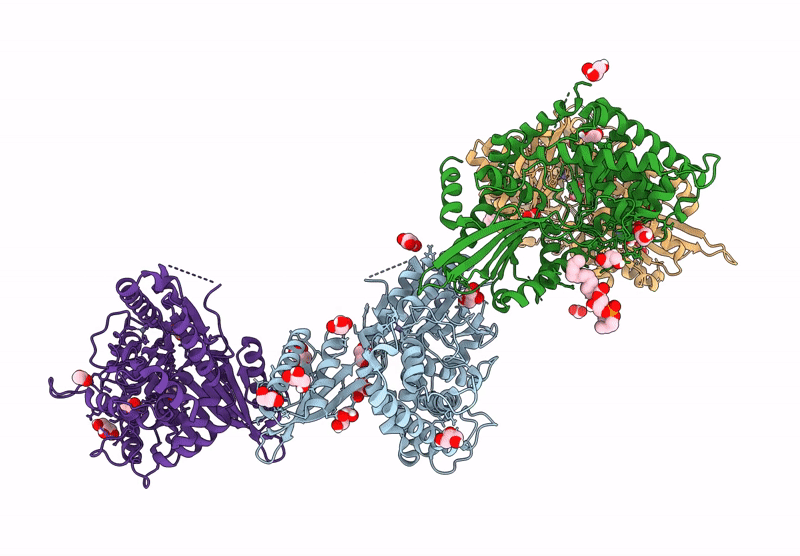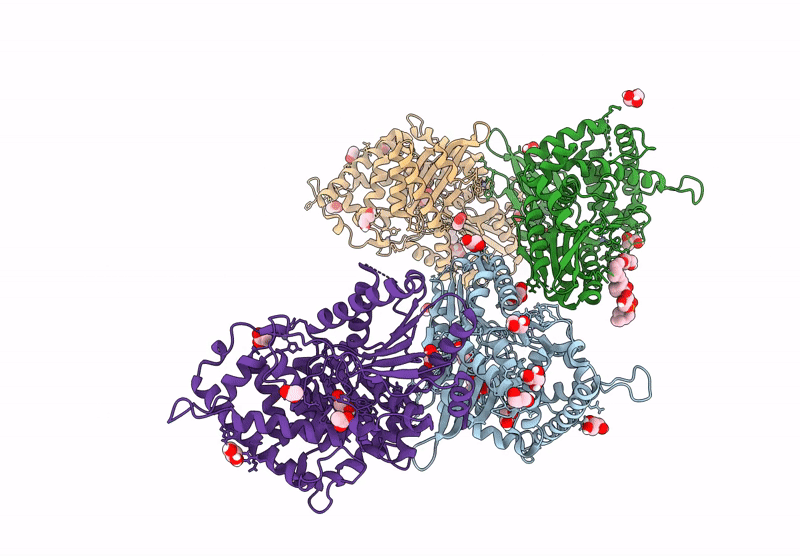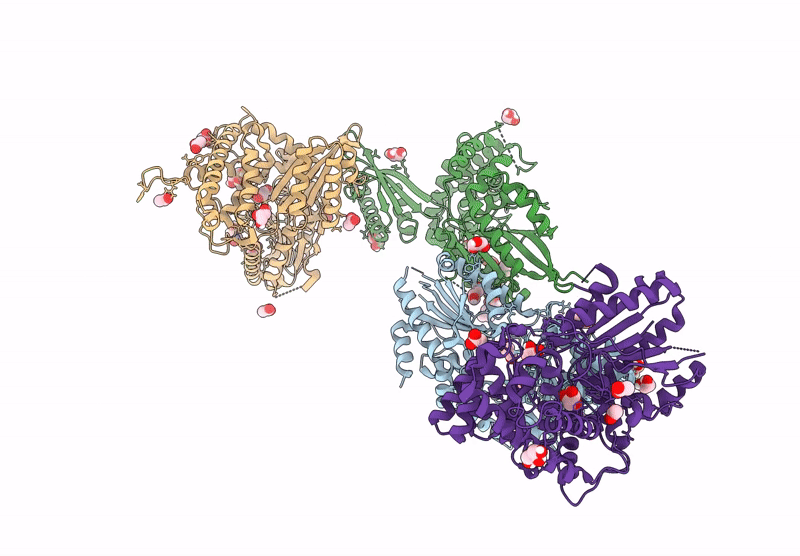
Crystal Structure Of The C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, ZN, LPC, ACT |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 In Complex With 1,5-Insp8 (Phosphatase Dead Mutant, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SPM, ORN, PUT, EDO, I8P, 1JW |
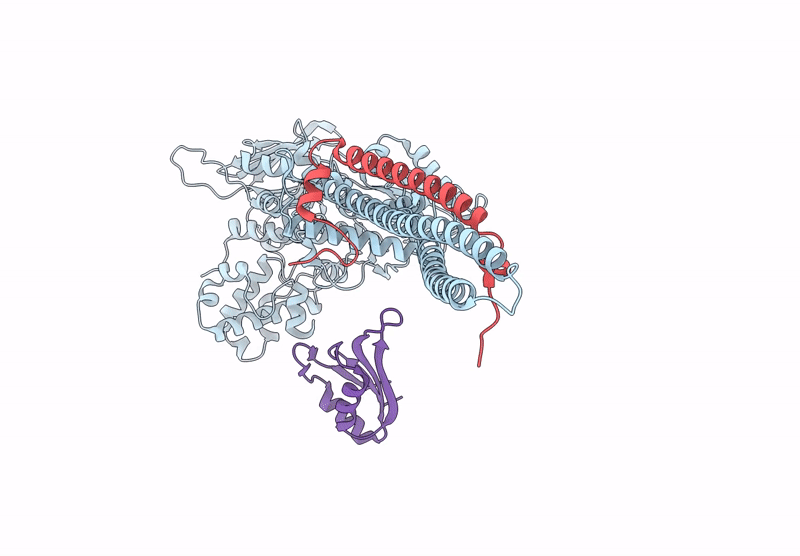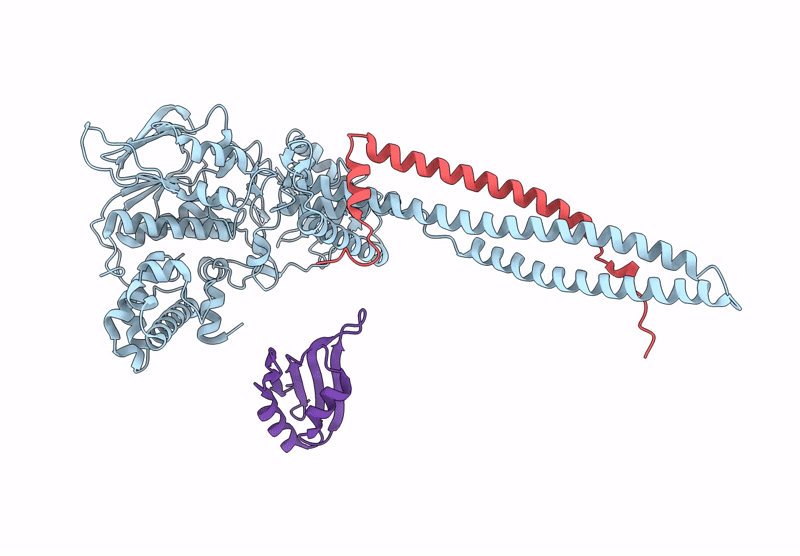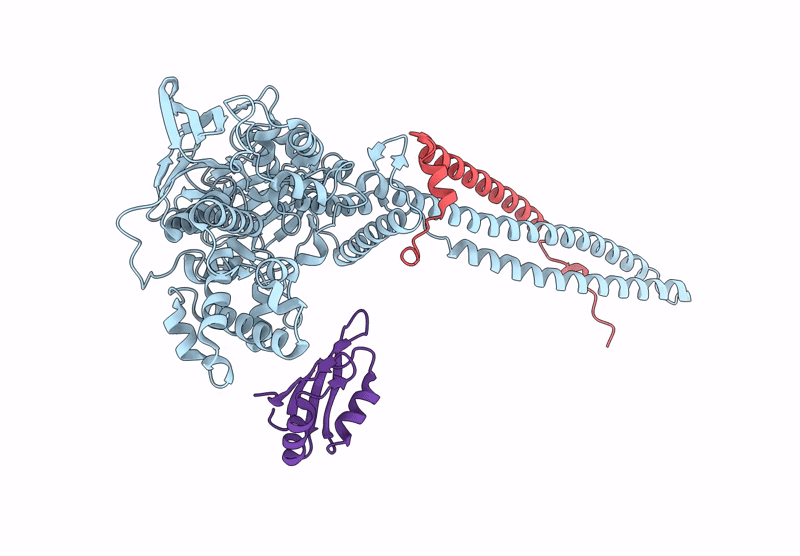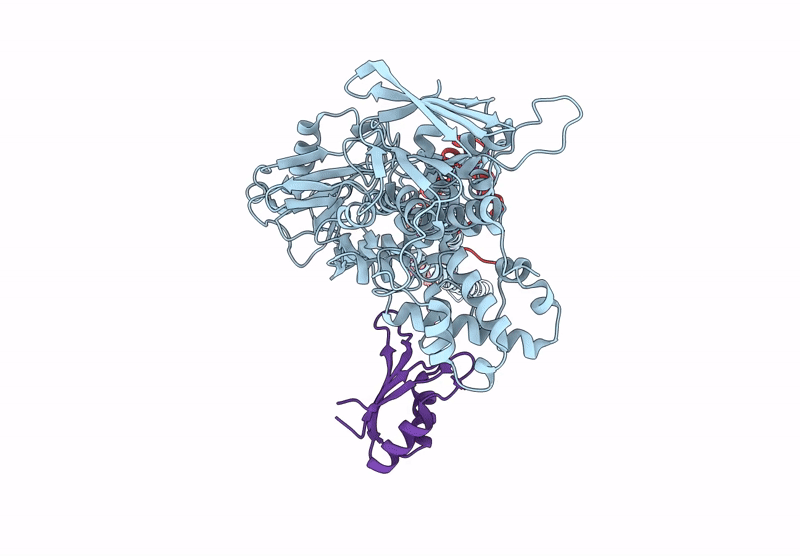 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: GENE REGULATION |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CL |
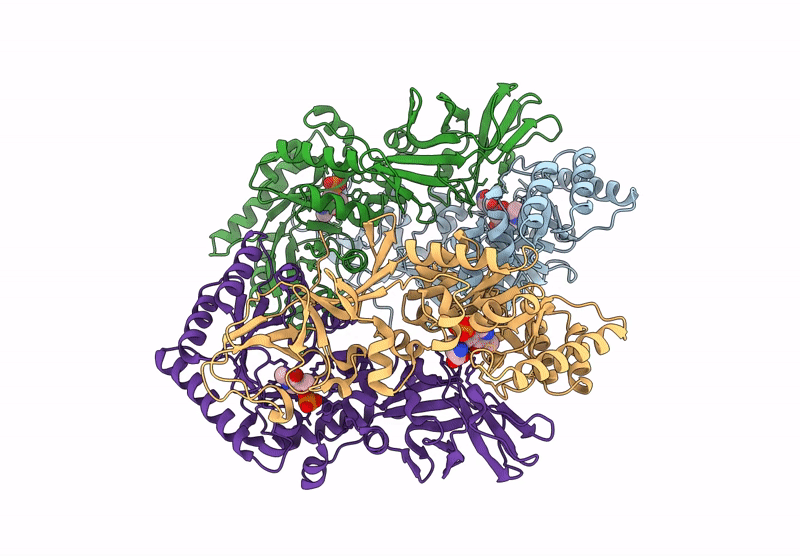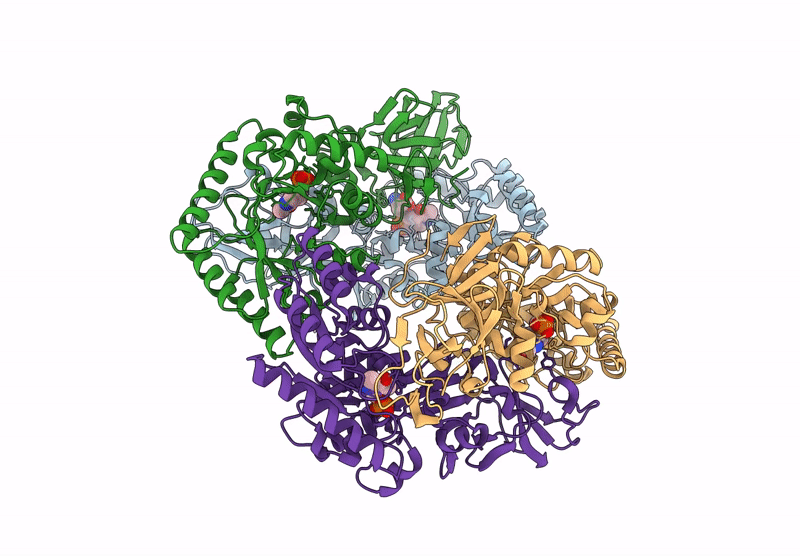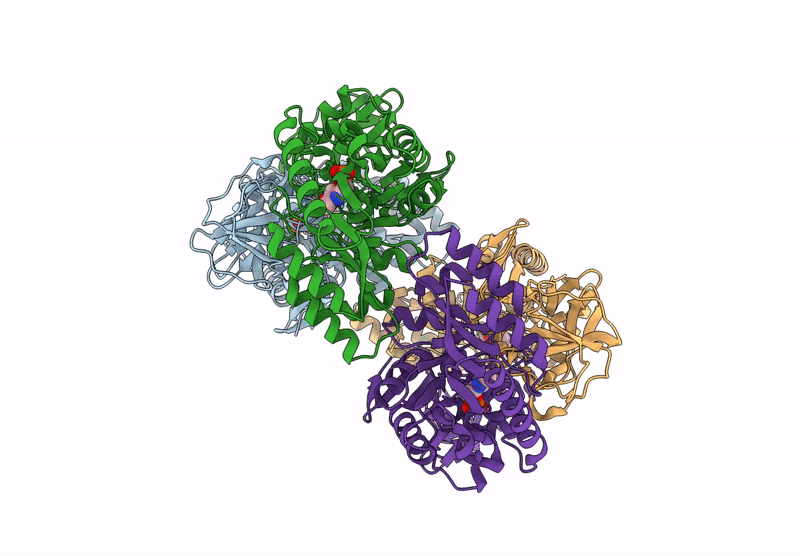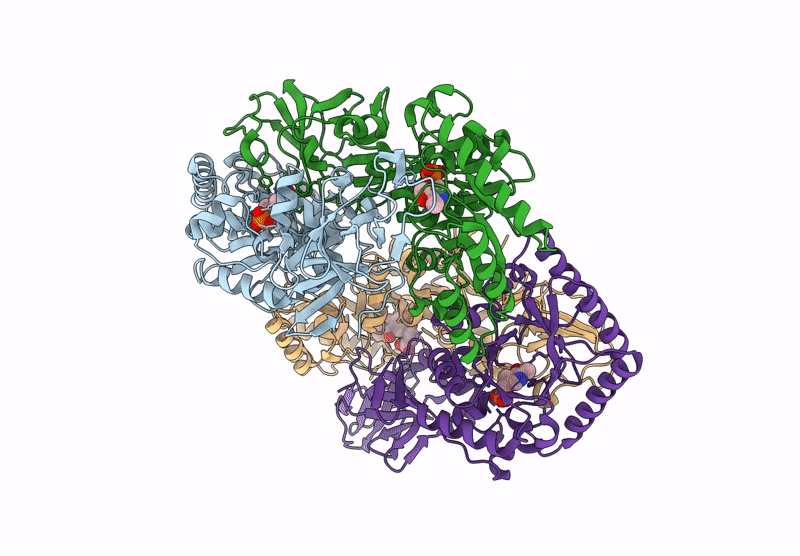 |
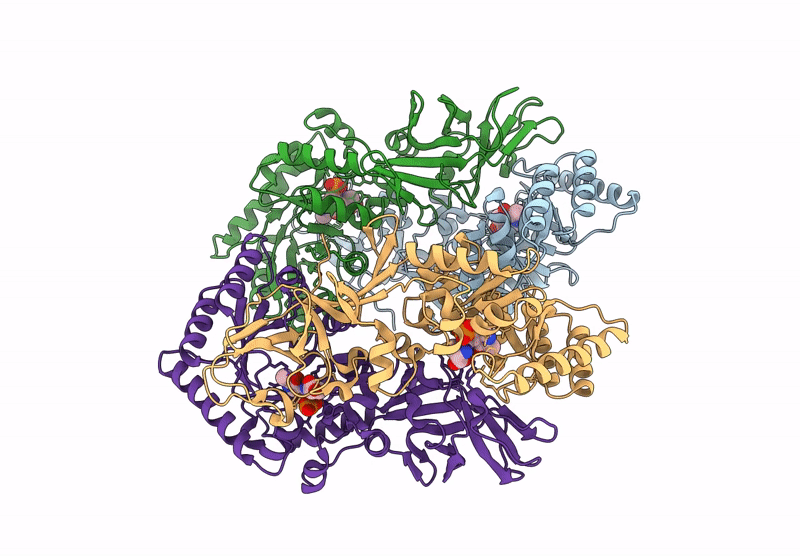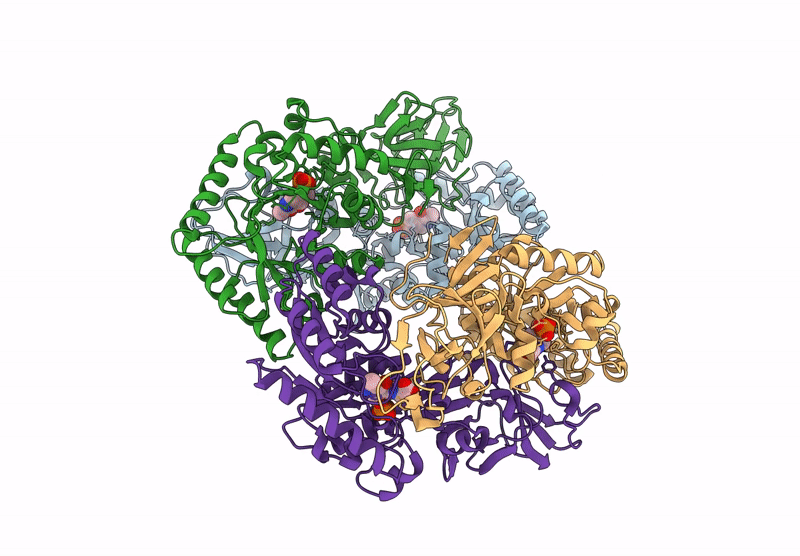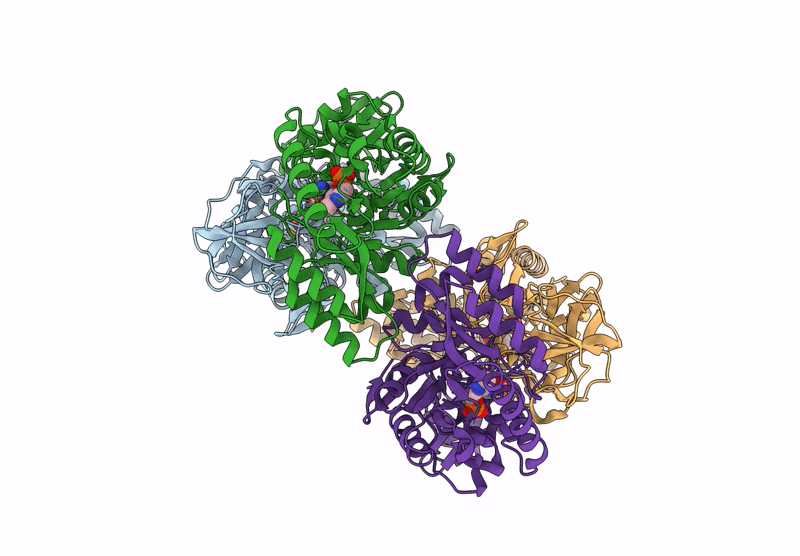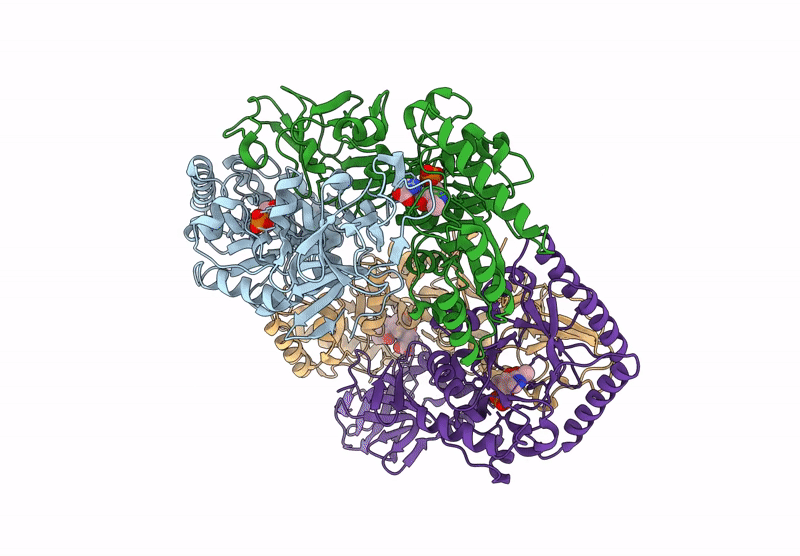
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
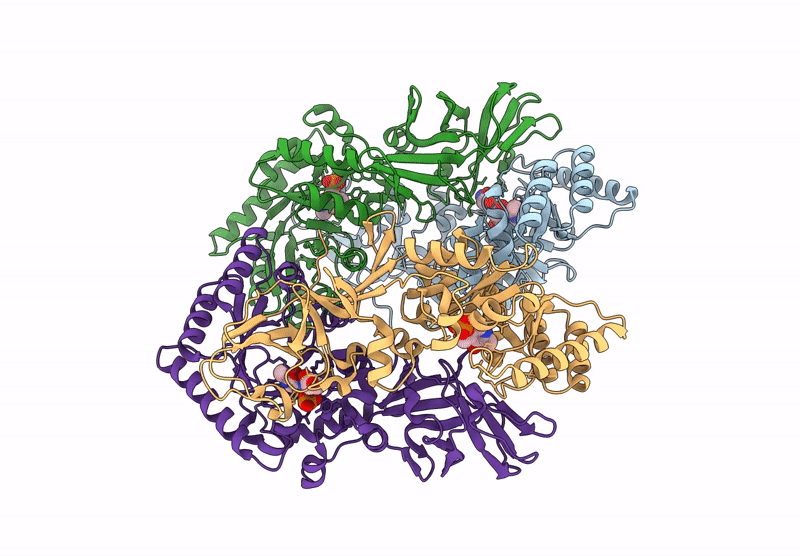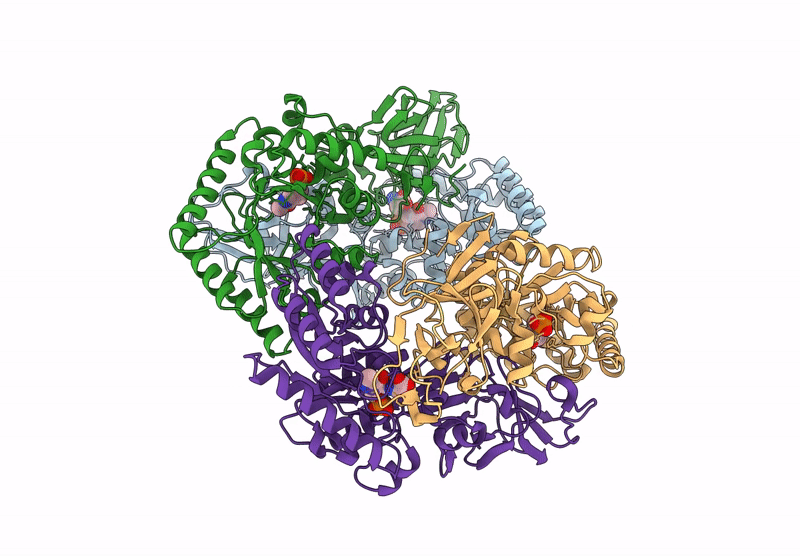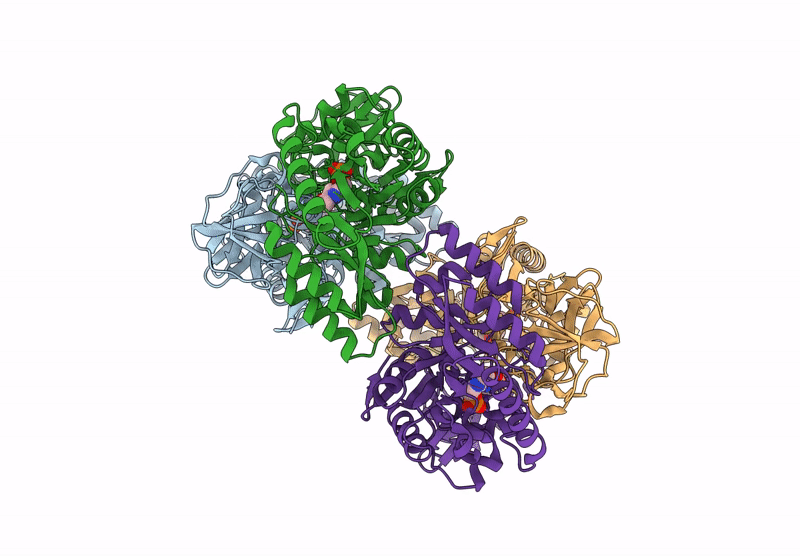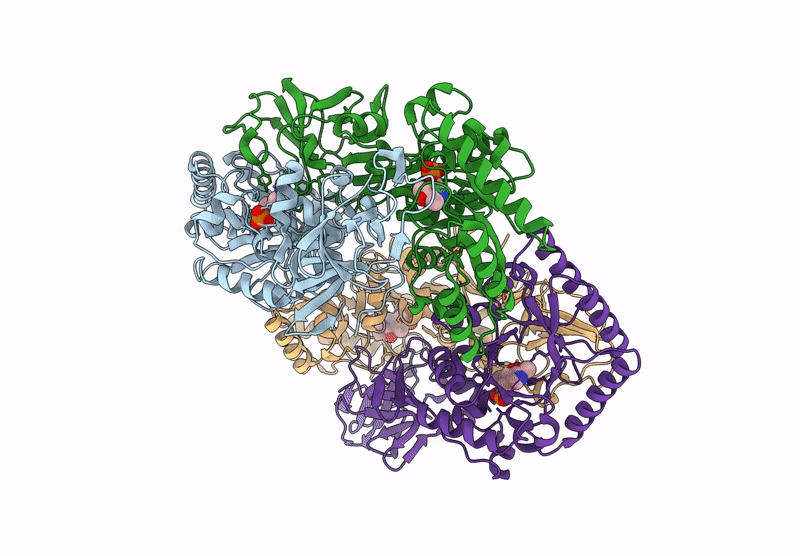
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, ALA, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
 |
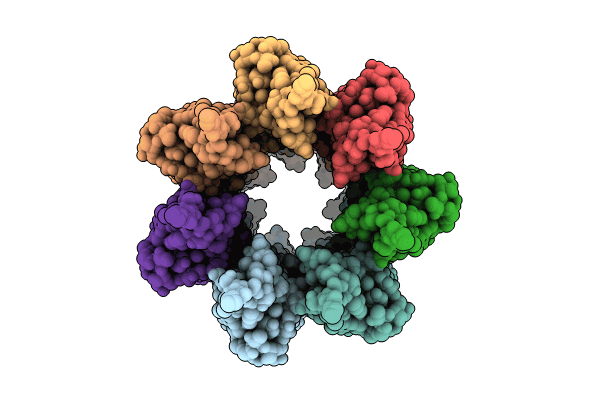
Cryoem Structure Of The Neck (1403-2314) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
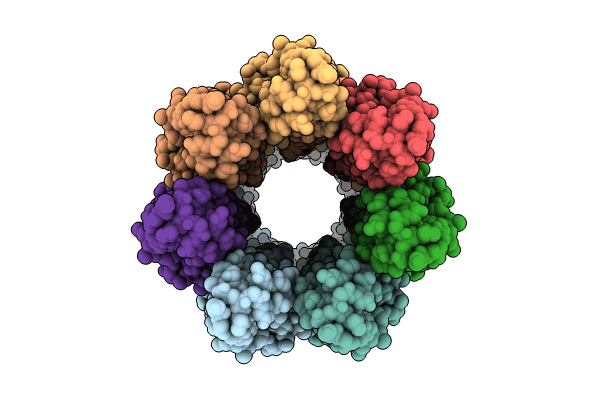
Cryoem Structure Of The Fragment-3 (2061-2397) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
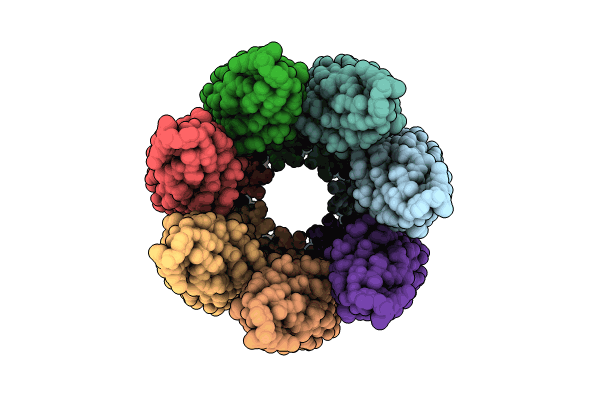
Cryoem Structure Of The Fragment-1 (3402-3733) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
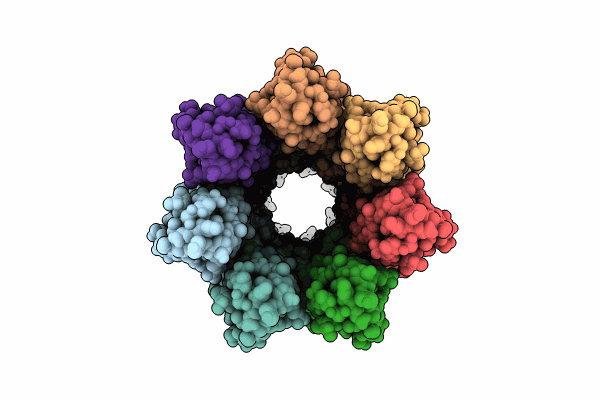
Cryoem Structure Of The Fragment-5 (2393-2807) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
Cryoem Structure Of The Fragment-6 (3571-4078) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
Cryoem Structure Of The Fragment-4 (4074-4421) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
Cryoem Structure Of The Base (4151-5009) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |
 |
Cryoem Structure Of The Fragment-2 (2892-3236) In The Grappling Hook Protein A (Ghpa) In The Bacterium Aureispira Sp. Ccb-Qb1
Organism: Aureispira sp. ccb-qb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CELL ADHESION |

