Search Count: 17
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-10-11 Classification: HYDROLASE Ligands: AMP, SO4 |
 |
Organism: Enterobacter aerogenes
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: CD, AMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: AMP, SO4, EDO |
 |
Structure Of The Conserved Yeast Listerin (Ltn1) N-Terminal Domain, Monoclinic Form
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-07-06 Classification: LIGASE Ligands: K, EDO |
 |
Structure Of The Conserved Yeast Listerin (Ltn1) Selenomethionine-Substituted N-Terminal Domain, Trigonal Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-07-06 Classification: LIGASE Ligands: K |
 |
Organism: Salmonella phage spc32h
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Salmonella phage spc32h
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2013-06-12 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-08-22 Classification: Transferase/Transferase Inhibitor Ligands: LNH, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-11-01 Classification: HYDROLASE Ligands: BOG, SO4 |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-04-08 Classification: GENE REGULATION Ligands: ZN, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-04-08 Classification: GENE REGULATION Ligands: ZN, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-04-08 Classification: GENE REGULATION Ligands: ZN, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2003-04-08 Classification: GENE REGULATION Ligands: ZN, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2003-04-08 Classification: GENE REGULATION Ligands: ZN, OAD |
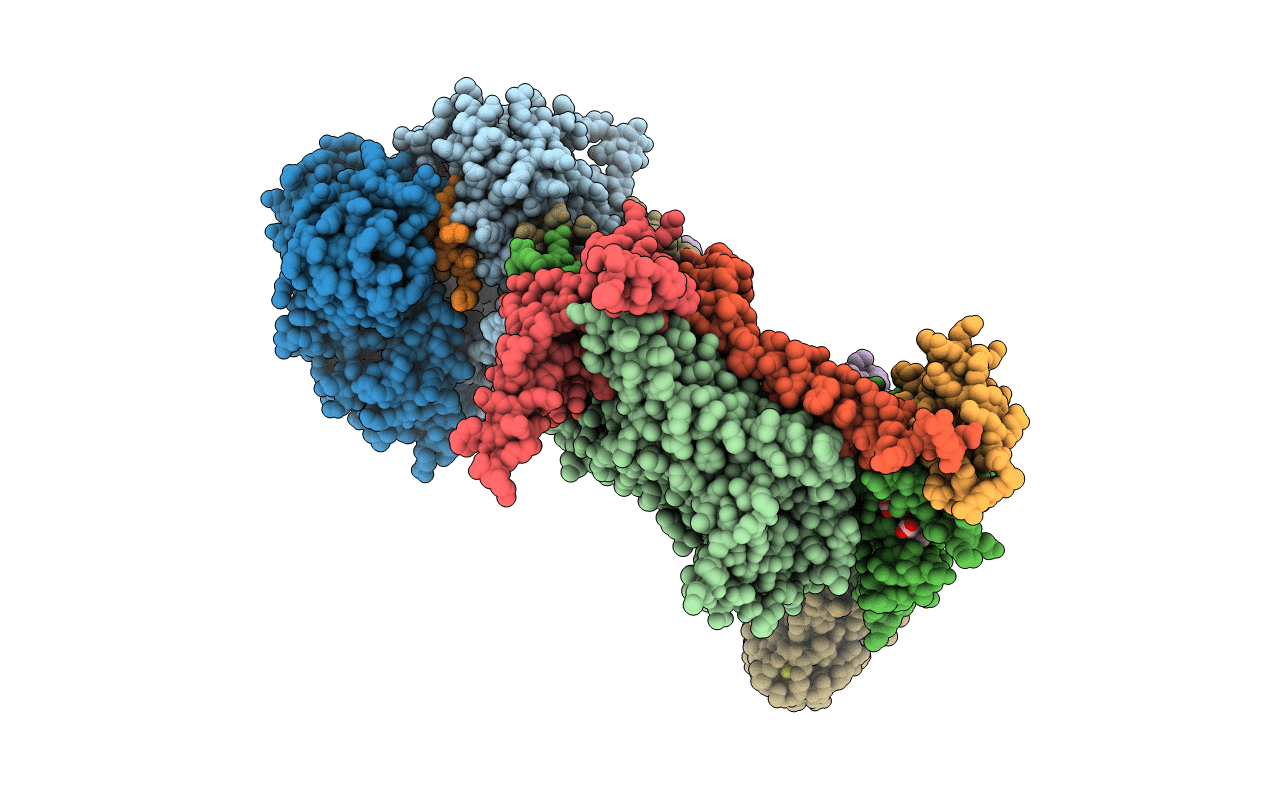 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1999-01-13 Classification: ELECTRON TRANSPORT Ligands: HEM, HEC, FES |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1999-01-06 Classification: ELECTRON TRANSPORT Ligands: HEM, HEC, FES |

