Search Count: 1,664
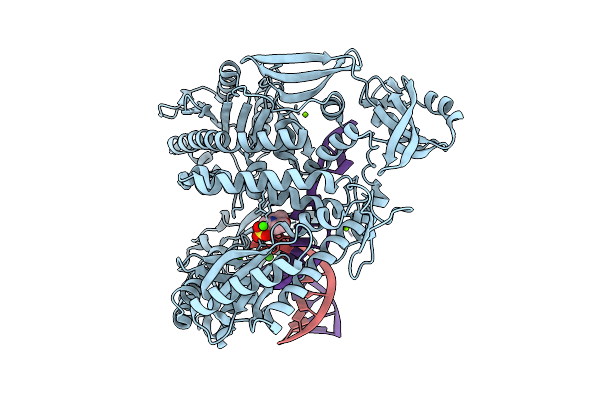 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: CA, MG, MES |
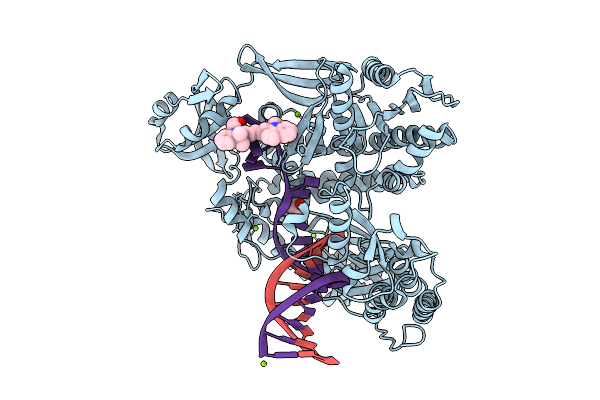 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG, 5CY, GOL |
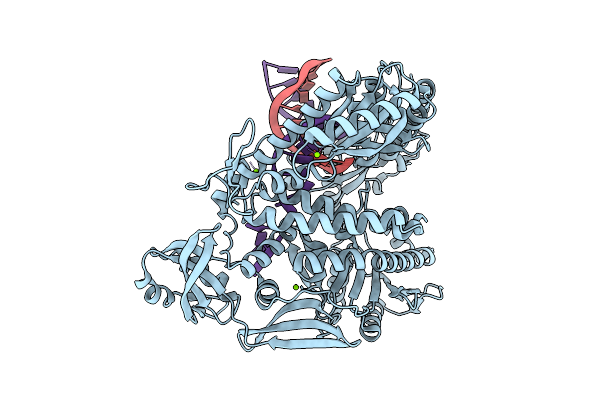 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
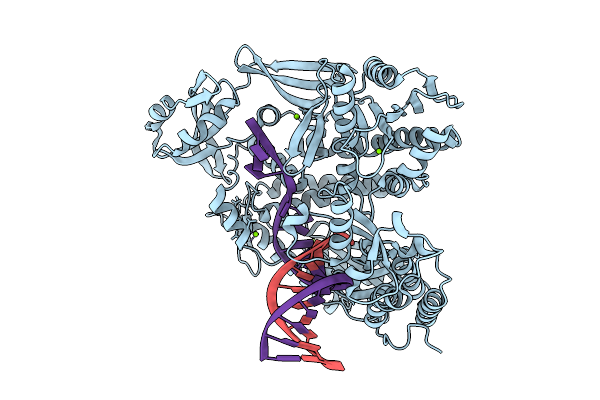 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
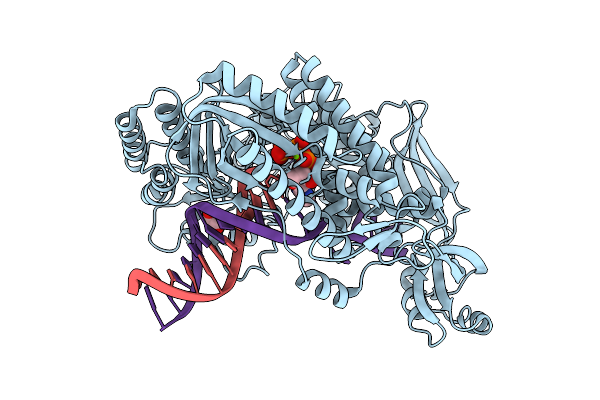 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, EDO |
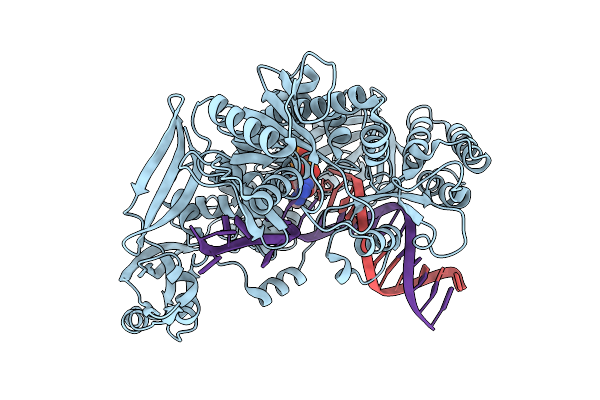 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, CL |
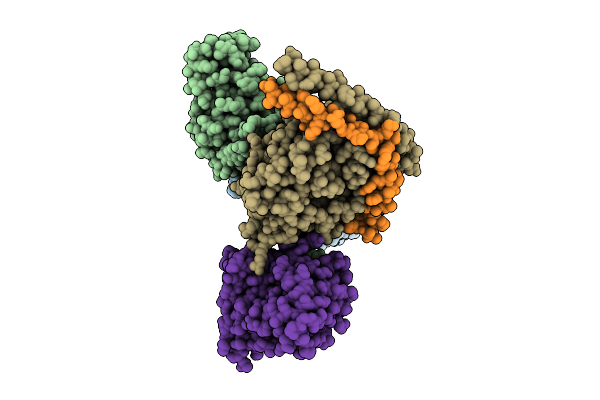 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
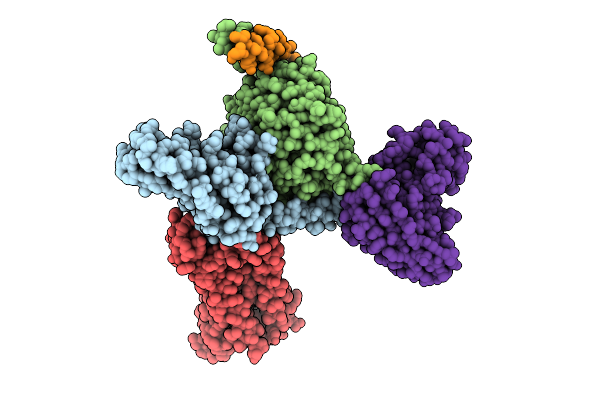 |
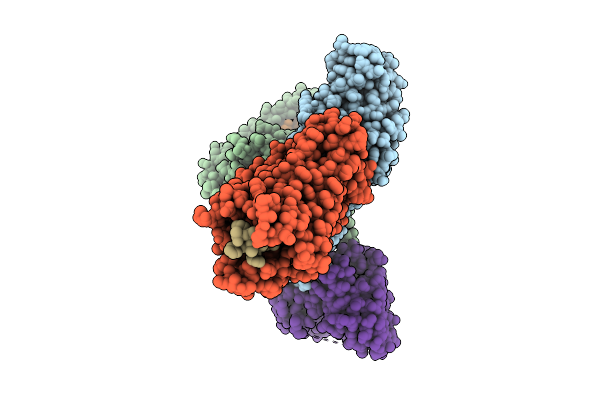
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN Ligands: A1D9A |
 |
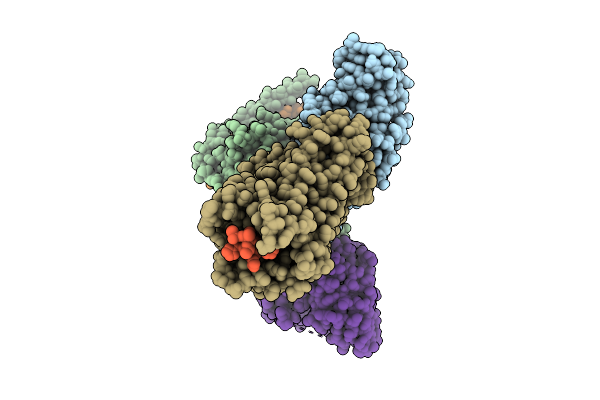
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
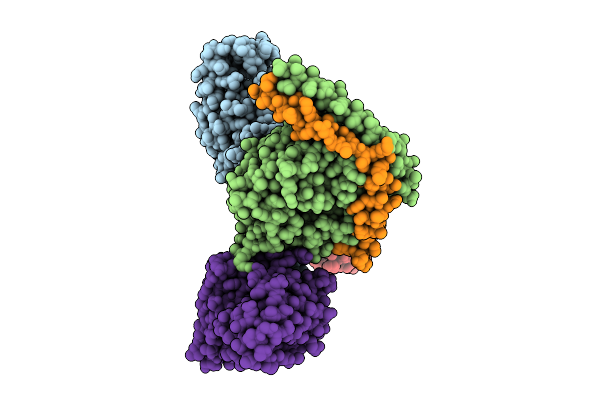
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
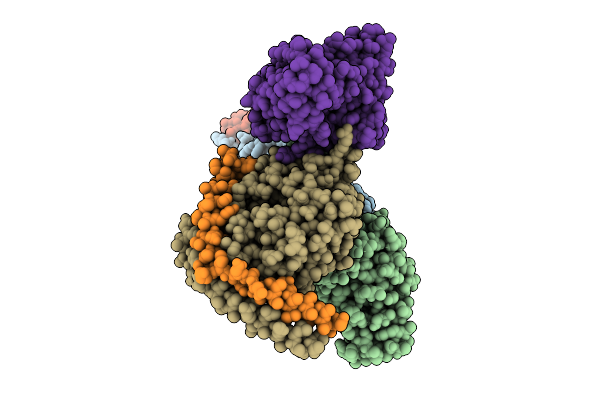
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN Ligands: A1L6Y |
 |
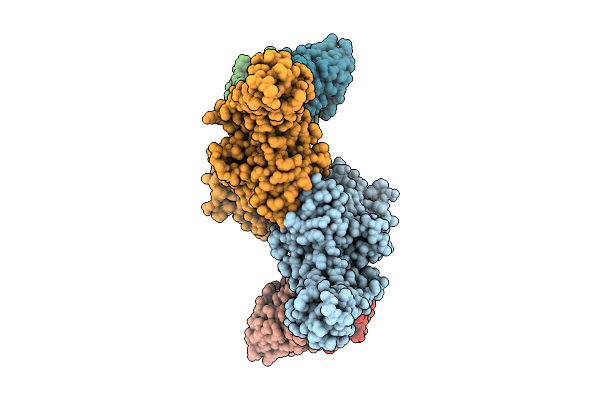
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
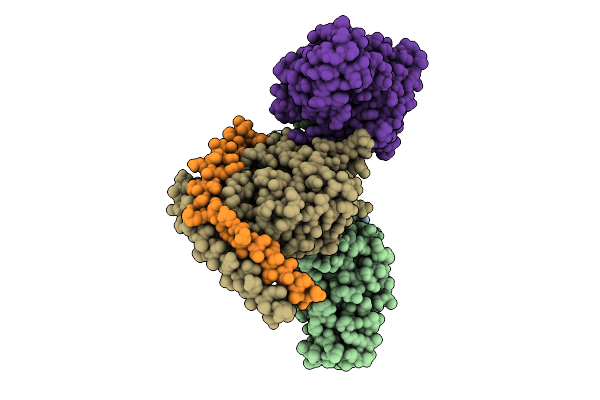 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
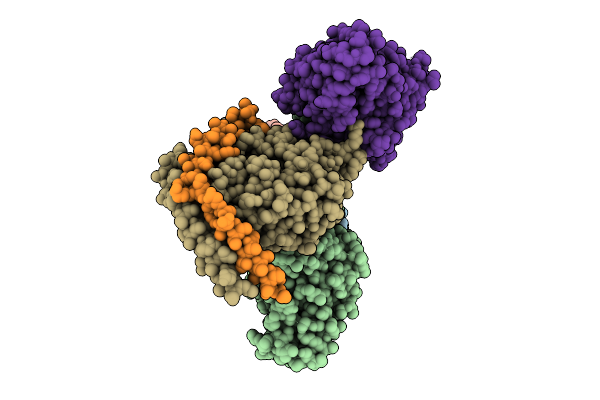 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
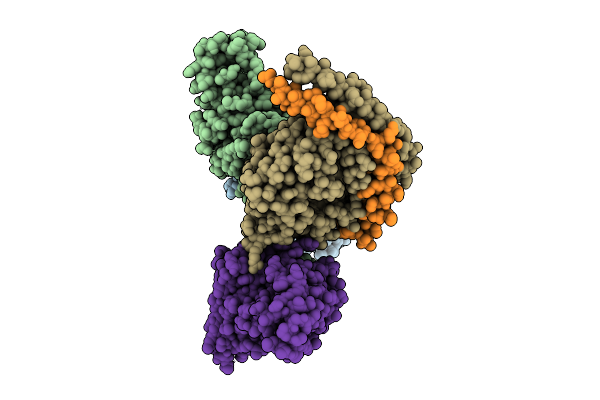 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
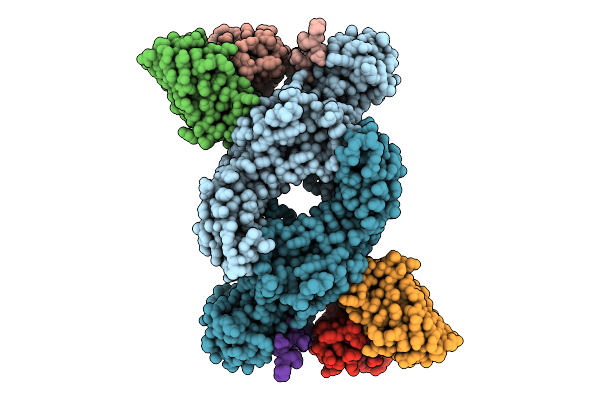
Organism: Bos taurus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
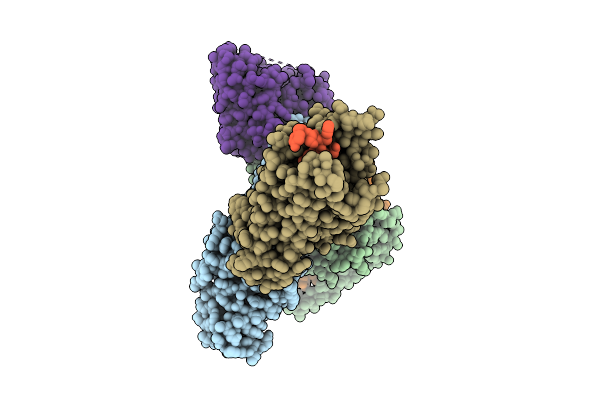
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
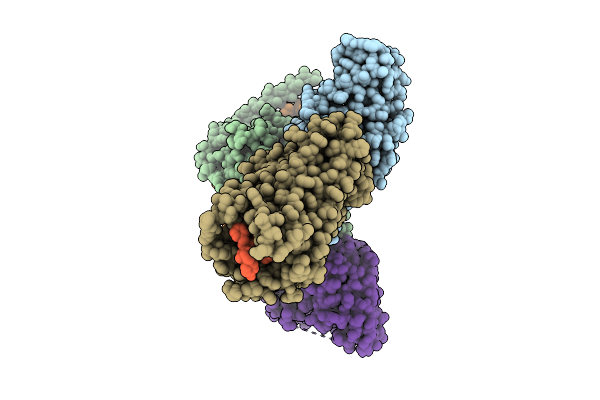
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
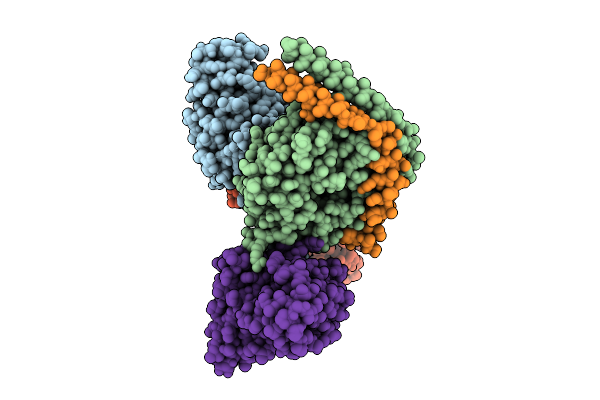
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: SIGNALING PROTEIN |

