Search Count: 128
 |
Organism: Escherichia coli, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: HYDROLASE |
 |
Organism: Escherichia coli, Gallus gallus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: TRS, GOL, BCN |
 |
Crystal Structure Of Trimeric K2-2 Tsp In Complex With Tetrasaccharide And Octasaccharide
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: ACE, EDO |
 |
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-07-19 Classification: ELECTRON TRANSPORT Ligands: HEC, FC6 |
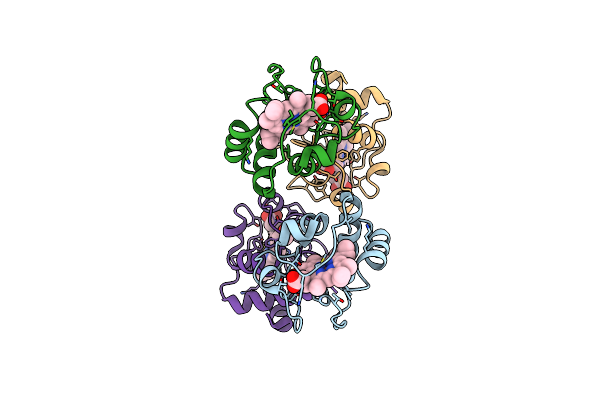 |
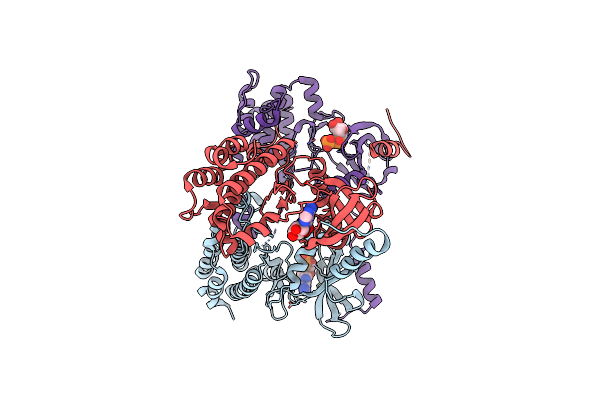
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-07-19 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-05-31 Classification: CYTOSOLIC PROTEIN Ligands: ADP |
 |
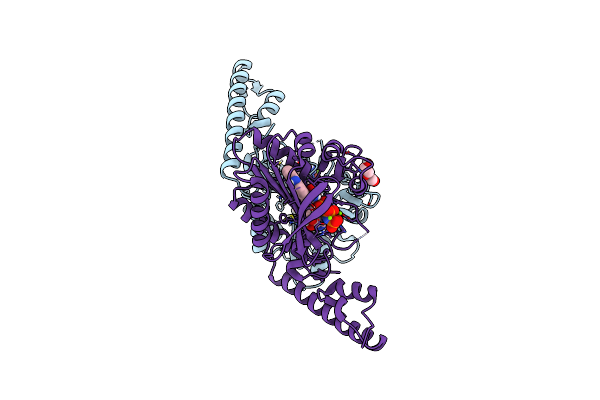
Complex Structure Of Catalytic, Small, And A Partial Electron Transfer Subunits From Burkholderia Cepacia Fad Glucose Dehydrogenase
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, HEM |
 |
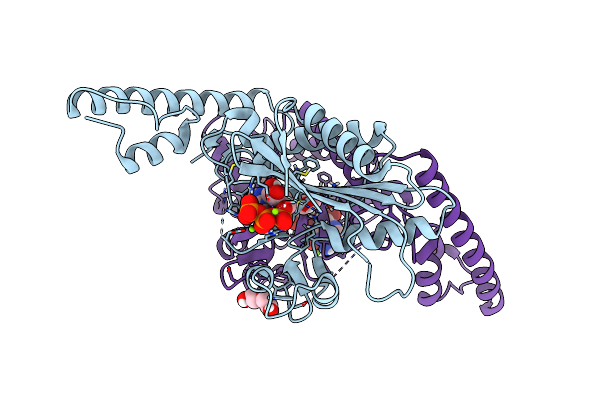
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Amppnp And Acridone
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ANP, T7Q, MG, GOL, PEG |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Atp And Acridone After 24 Hours Of Crystal Growth
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, PEG, MG, GOL |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Rs1) Bound To Atp And Acridone After 2- Weeks Of Crystal Growth
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, PG4, MG, GOL |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For Acridone Amino Acid (Ast) Bound To Atp And Acridone
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-07 Classification: LIGASE Ligands: ATP, T7Q, MG, GOL, AMP, PEG |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A P1 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: LMR, IMD, GOL |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A C2 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: CIT, CO3 |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In Complex With Products
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: IMD, GOL, 98X |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-04-13 Classification: LIGASE Ligands: MG, ATP |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-13 Classification: HYDROLASE |
 |
Organism: Capsicum annuum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: HEC, FC6 |

