Search Count: 135
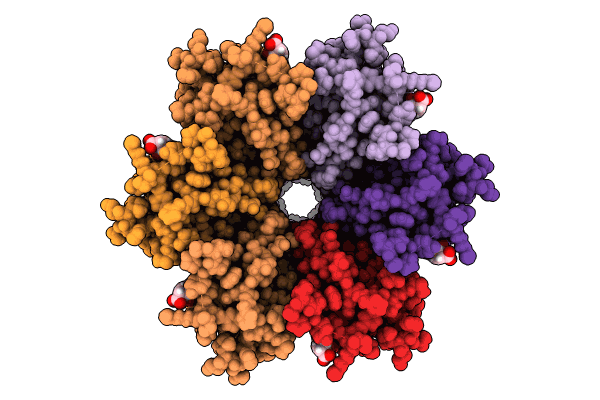 |
Human Cx36/Gjd2 Gap Junction Channel With Pore-Lining N-Terminal Helices In Porcine Brain Lipids.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, Y01 |
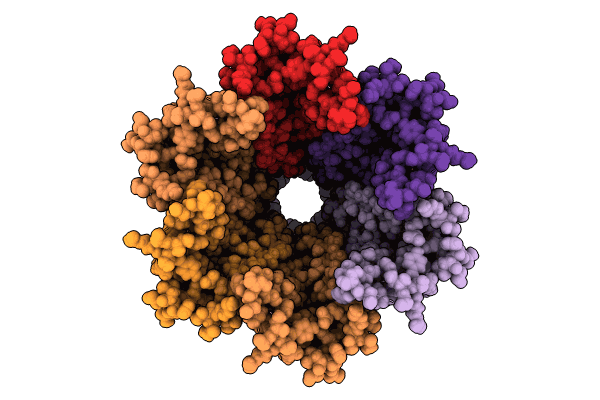 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
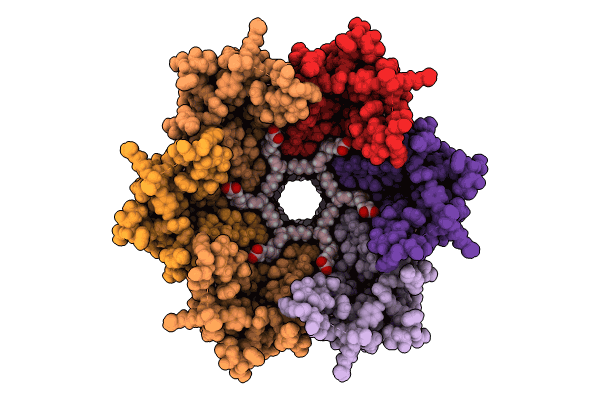 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, ACD |
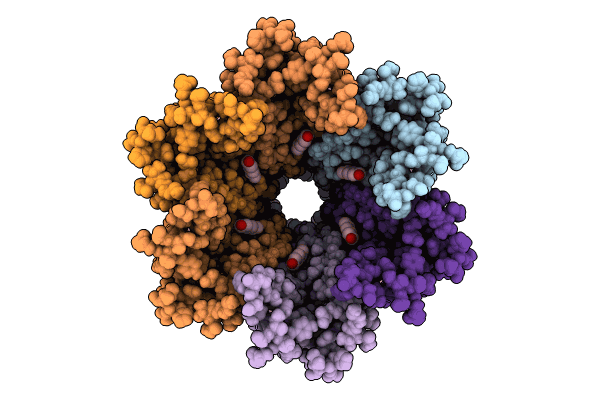 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, HE2 |
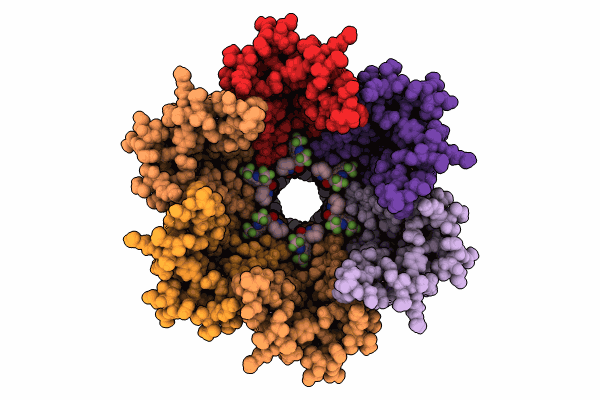 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: YMZ, MC3 |
 |
Human Cx36/Gjd2 (Ala14-Deleted Mutant) Gap Junction Channel In Porcine Brain Lipids
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Human Cx36/Gjd2 (Ala14 Deletion Mutant) Gap Junction Channel Prepared With Mefloquine, Showing No Bound Mefloquine
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
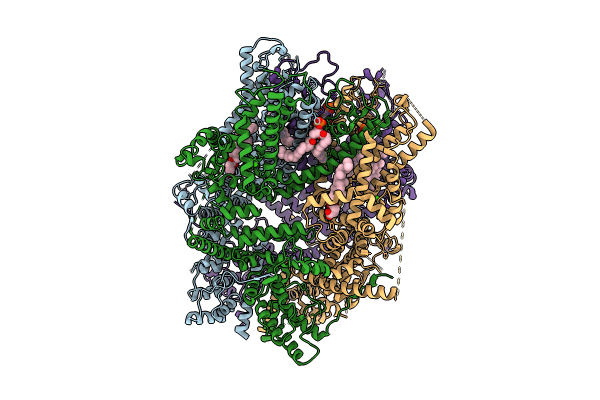 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: LPP, ZN, Y01, CA |
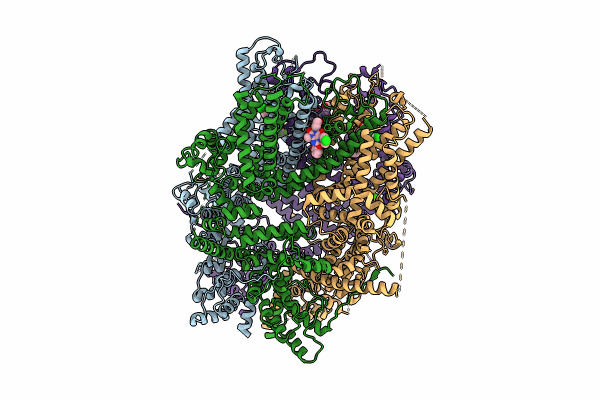 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: PJQ, ZN, CA |
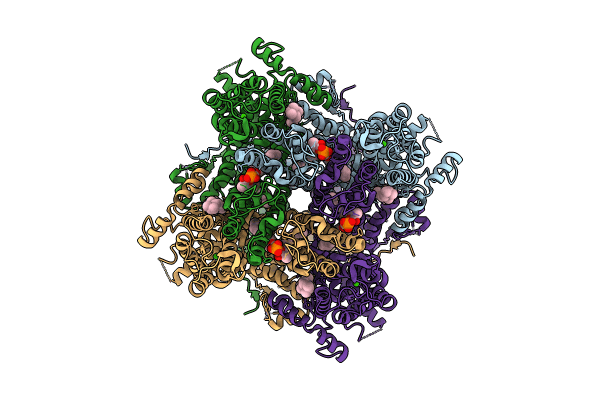 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: Y01, ZN, CA, LPP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.49 Å Release Date: 2024-09-18 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In High Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, 1PE |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In Low Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, S8I, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, PGE, S7L |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE |
 |
Human Cx36/Gjd2 (N-Terminal Deletion Mutant) Gap Junction Channel In Soybean Lipids (D6 Symmetry)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Methylorubrum extorquens am1
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: W, MGD, FES, SF4, FMN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2023-07-19 Classification: SIGNALING PROTEIN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-07-19 Classification: SIGNALING PROTEIN Ligands: SO4 |

