Search Count: 957
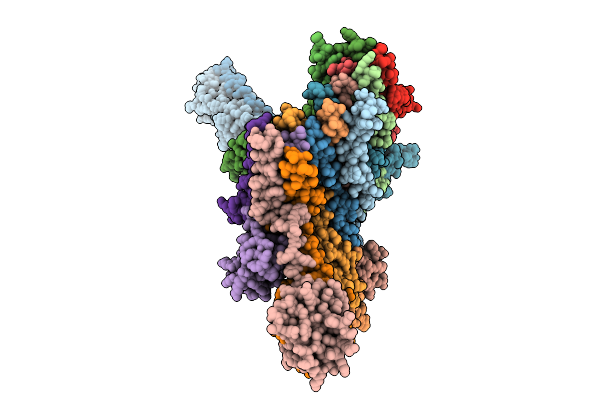 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Lacking The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
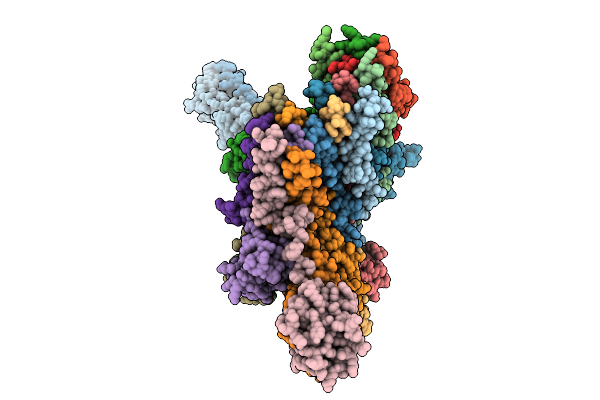 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Including The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
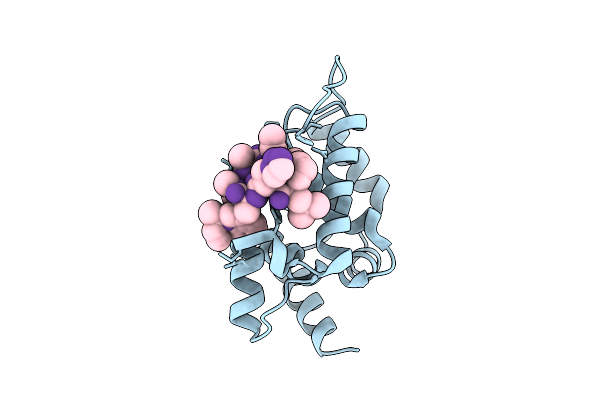 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
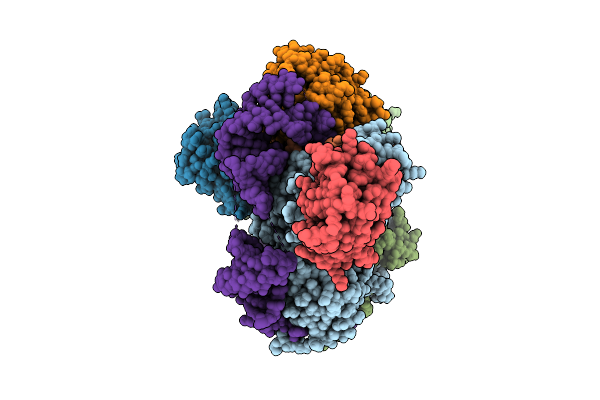 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Rhinovirus b14
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: XNV |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: CA, YZY, Y01, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: PTY, POV, Y01, ZN, CA, YZY |
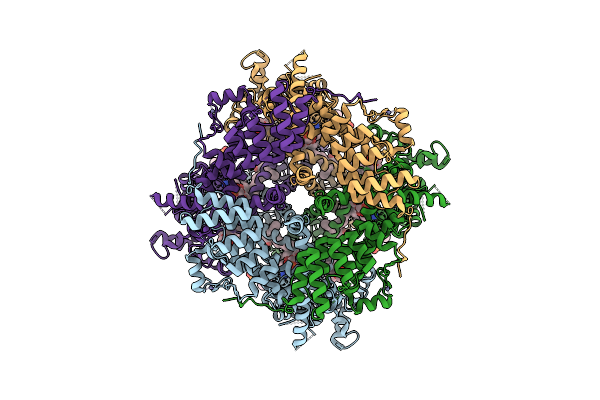 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: PTY, POV, Y01, ZN, CA, YZY |
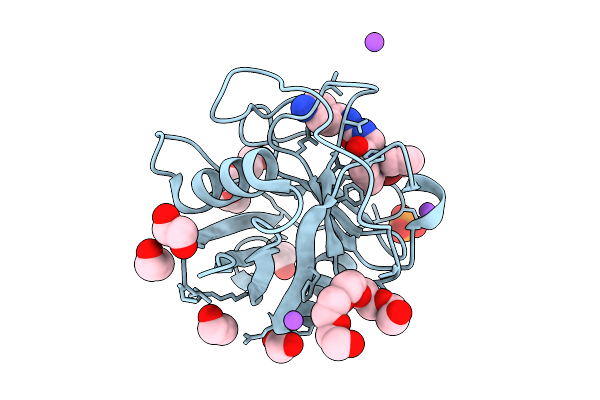 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: 9CK, EDO, 1PE, PGE, PO4, NA |
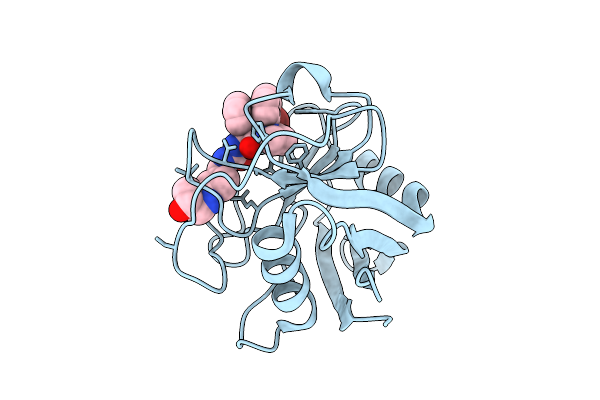 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: A1IU3 |
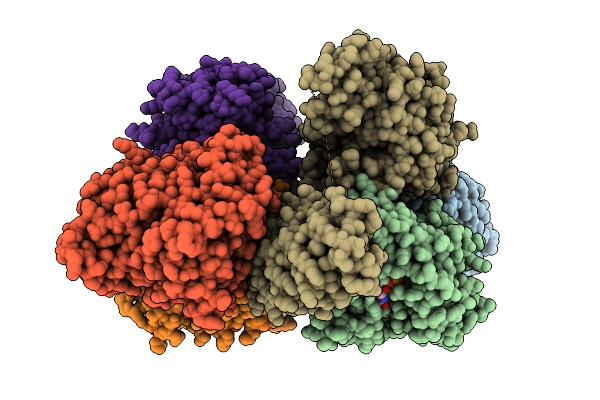 |
Organism: Homo sapiens, Synthetic construct, Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MOTOR PROTEIN Ligands: GTP, MG, EDO, GDP, ANP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: FUL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA Ligands: A1L8R |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp S759A Mutant In Complex With 20-40Mer Rna Incorporating Remdesivir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp Wild-Type In Complex With 20-40Mer Rna Incorporating Remdesivir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp S759A Mutant In Complex With 20-40Mer Rna Incorporating Atp
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |

