Search Count: 27
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Acp-Bound State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Clbi Acp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
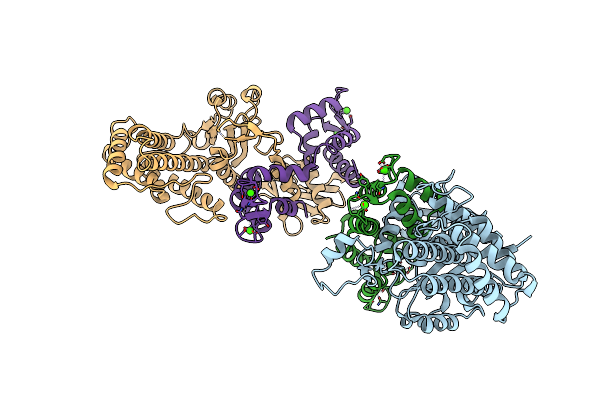
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Its Precursor Module, Clbh
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
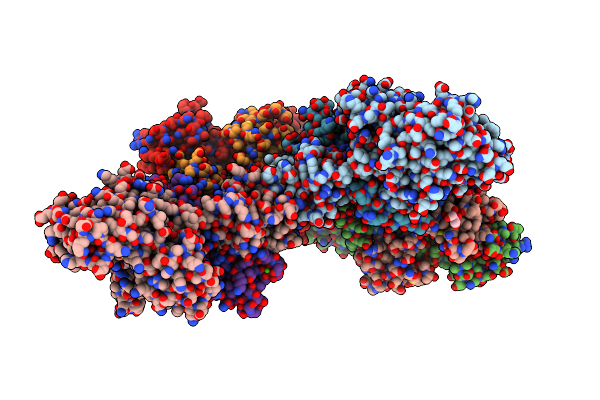
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
 |
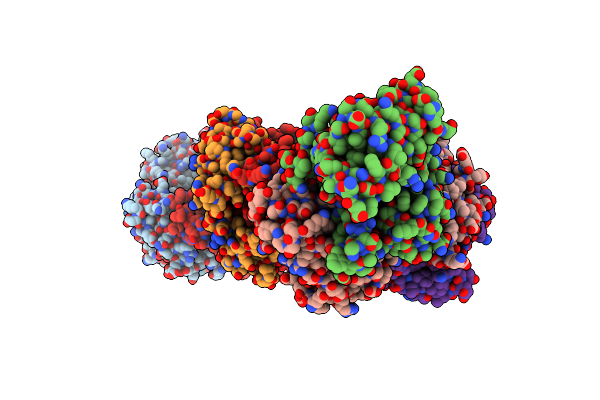
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |
 |
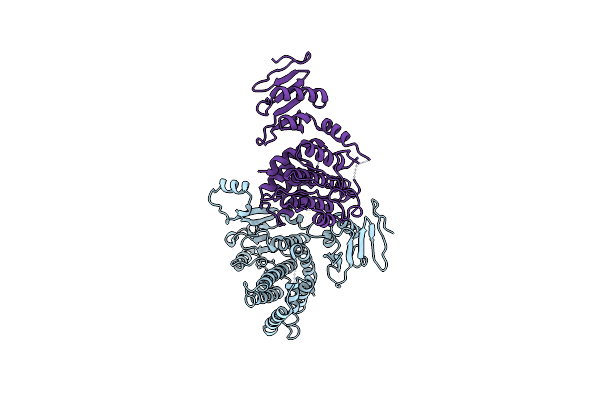
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Free Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: MG |
 |
Crystal Structure Of The Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Crystal Structure Of The Chicken Toll-Like Receptor 15 Tir Domain (2-Mercaptoethanol Adduct)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-31 Classification: IMMUNE SYSTEM Ligands: SO4, GOL |
 |
Crystal Structure Of The Chicken Toll-Like Receptor 15 Tir Domain (Glutathione Adduct)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-31 Classification: IMMUNE SYSTEM Ligands: GSH |
 |
Crystal Structure Of A Complex Between The Antirepressor Gmar And The Transcriptional Repressor Mogr
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2022-11-23 Classification: PROTEIN BINDING/TRANSCRIPTION |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-11-09 Classification: PROTEIN BINDING |
 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-12-30 Classification: TRANSFERASE |
 |
Crystal Structure At 1.3 Angstroms Resolution Of A Novel Udg, Udgx, From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, PO4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: DU, SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |

