Search Count: 173
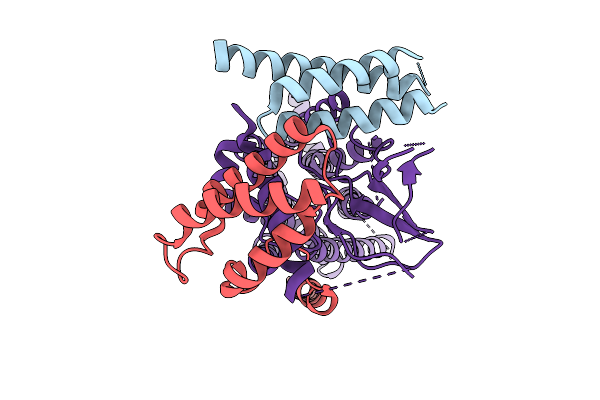 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
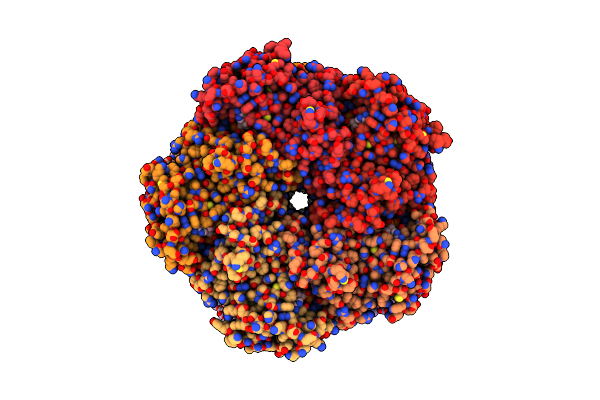 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: ATP, MG, GLN |
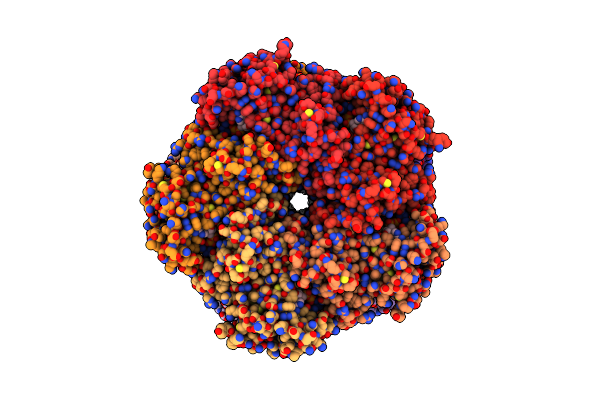 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: ATP, MG |
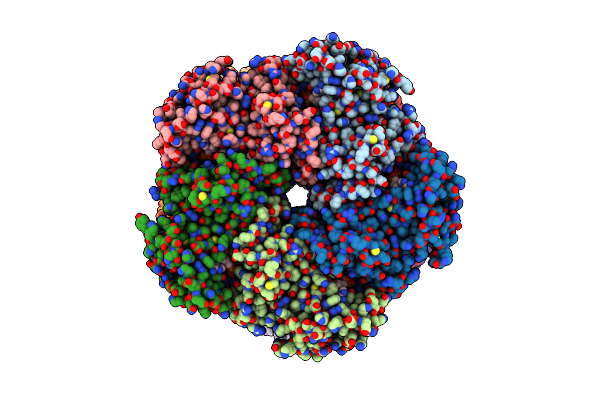 |
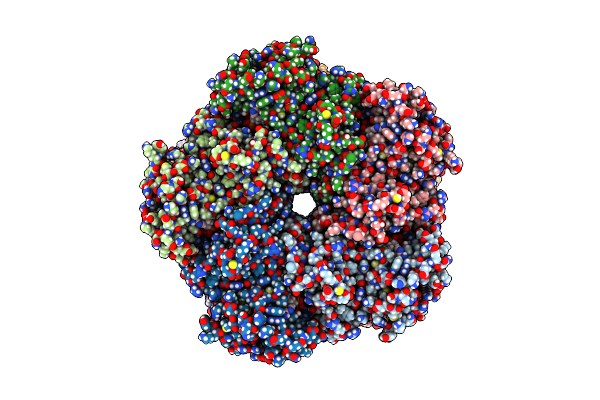
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: ADP, MG |
 |
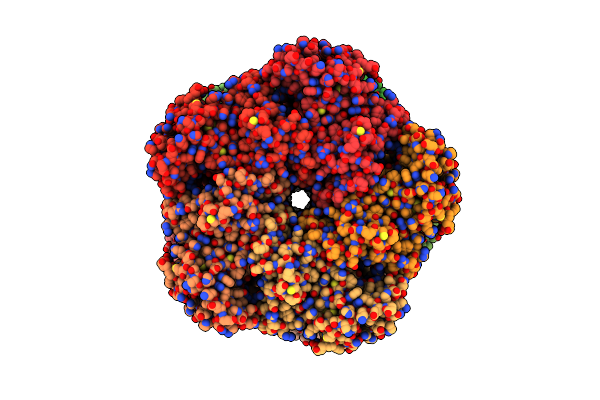
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: ADP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: MG |
 |
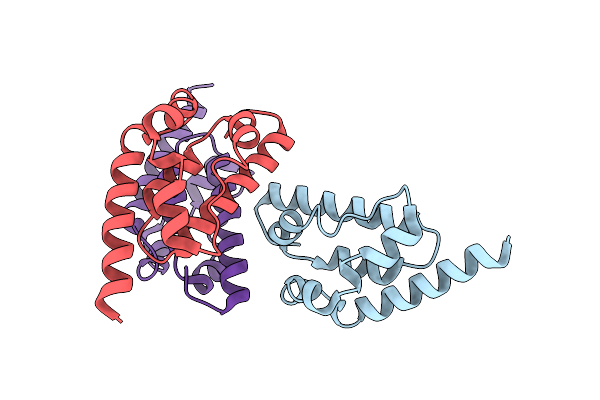
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: VIRAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ADP, ATP |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ACO, PO4 |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Acp-Bound State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Clbi Acp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Its Precursor Module, Clbh
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
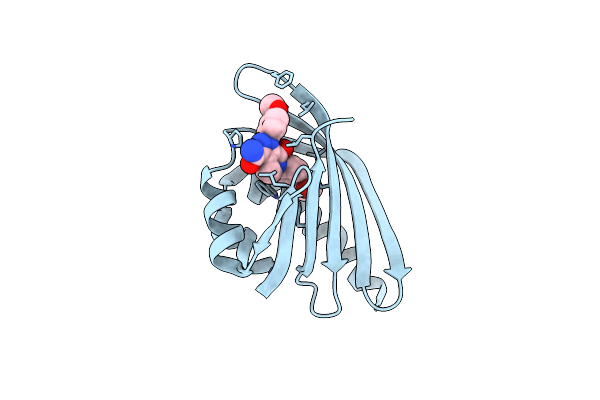
Crystal Structure Of Trehalose Synthase Mutant N253C From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-03-19 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-01-15 Classification: DE NOVO PROTEIN Ligands: GG2 |
 |
Crystal Structure Of Trehalose Synthase Mutant N253E From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase Mutant N253Q From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |

