Search Count: 37
 |
Organism: Bacillus subtilis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: HYDROLASE |
 |
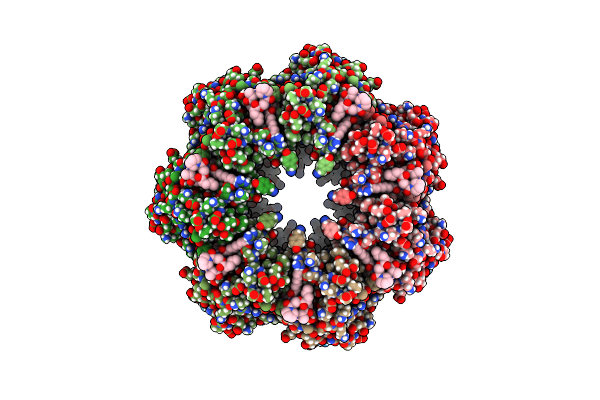
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: HYDROLASE |
 |
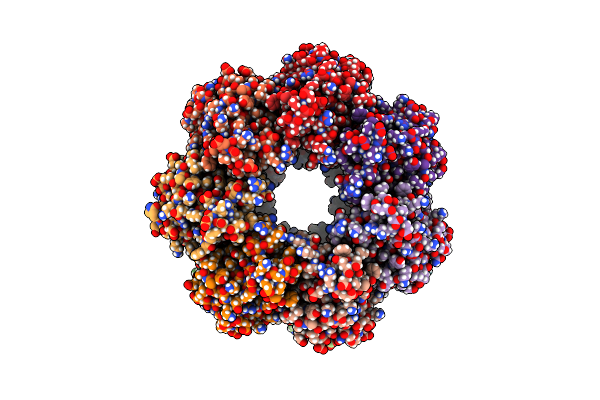
Organism: Bacillus subtilis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: HYDROLASE |
 |
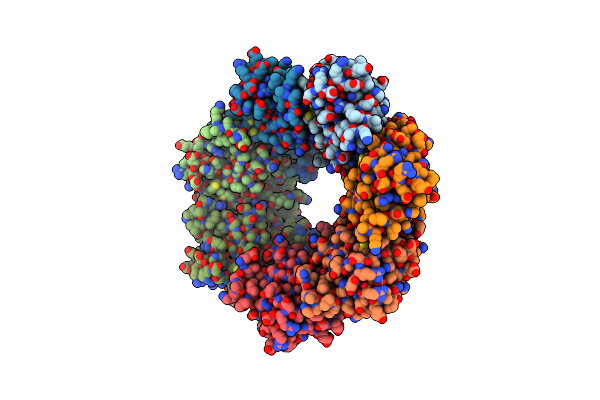
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: HYDROLASE |
 |
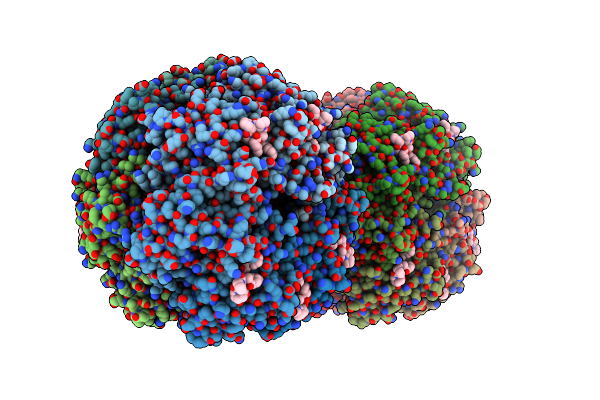
Crystal Structure Of Clpp From Bacillus Subtilis In Complex With Adep2 (Compressed State)
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-06-29 Classification: HYDROLASE |
 |
Crystal Structure Of Clpp From Bacillus Subtilis In Complex With Adep2 (Compact State)
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-06-29 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: CELL CYCLE Ligands: ADP, BEF, MG |
 |
Organism: Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: CELL CYCLE Ligands: ADP, BEF, MG |
 |
Crystal Structure Of Mouse Smc1-Smc3 Hinge Domain Containing A D574Y Mutation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-11-17 Classification: DNA BINDING PROTEIN |
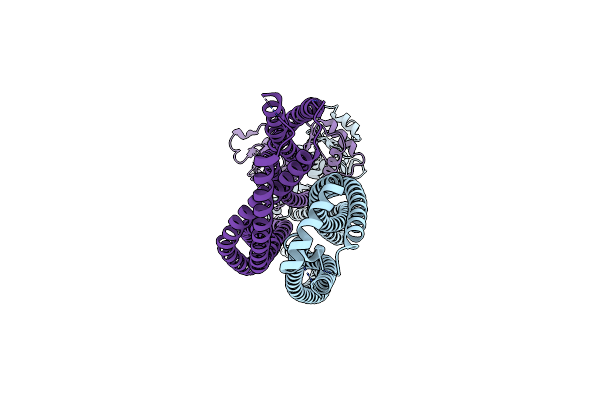 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: CELL CYCLE |
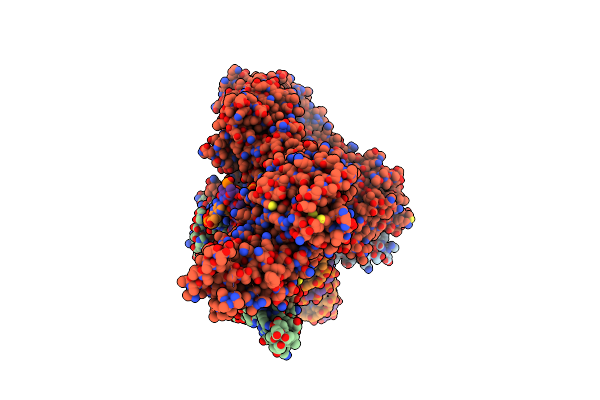 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-09-30 Classification: CELL CYCLE Ligands: ATP, MG |
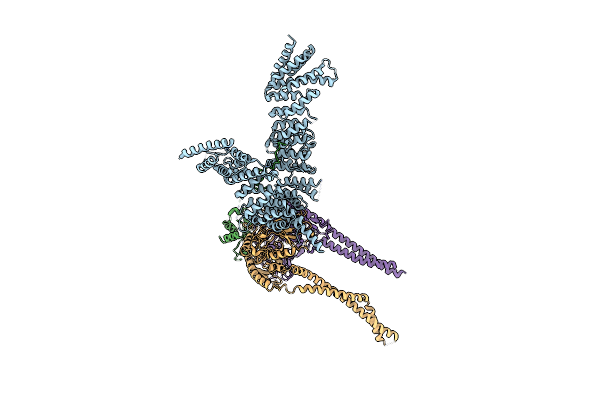 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: CELL CYCLE |
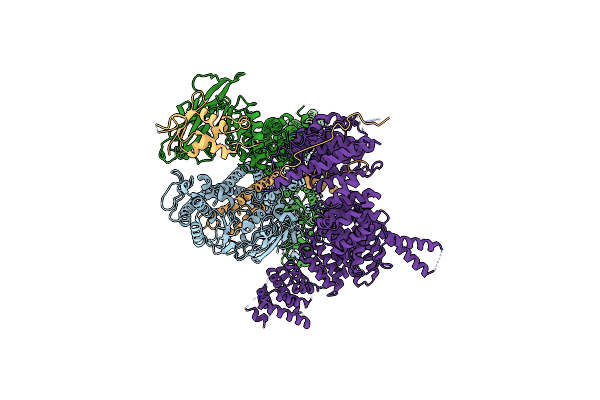 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: CELL CYCLE |
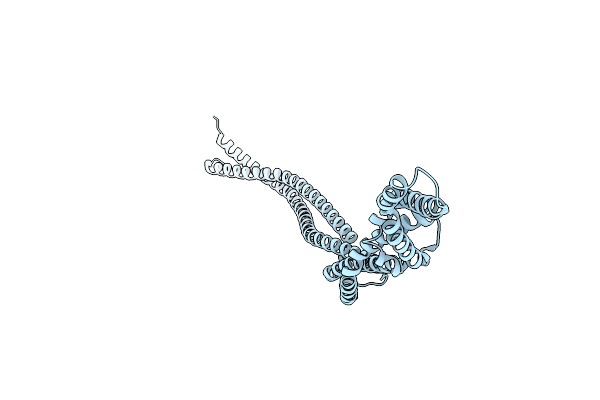 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-03-06 Classification: DNA BINDING PROTEIN |
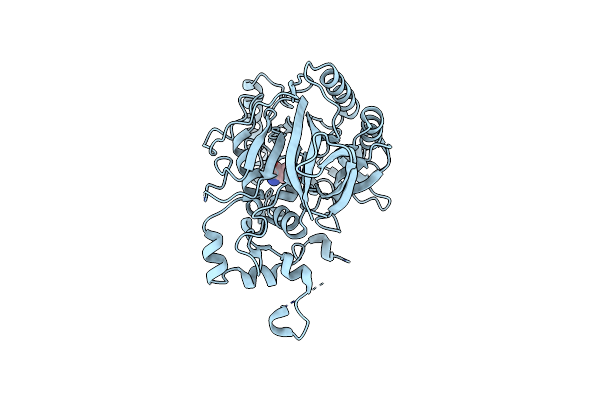 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: GLN |
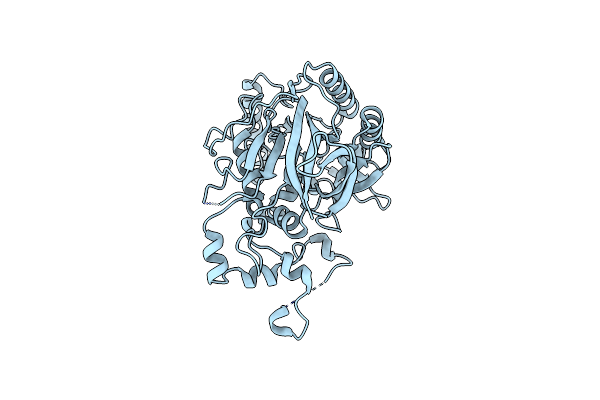 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-04-05 Classification: HYDROLASE |
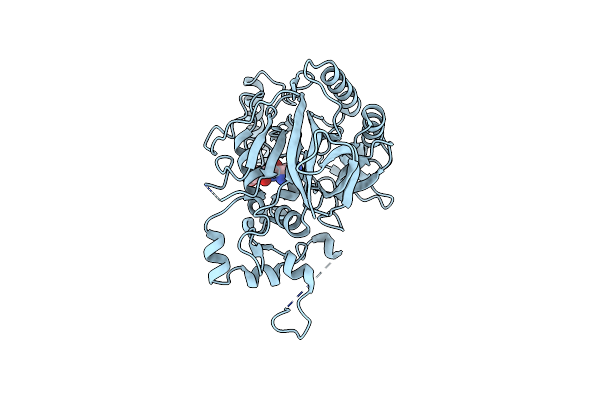 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d), Saccharomyces cerevisiae cen.pk113-7d
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2017-01-11 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-01-11 Classification: HYDROLASE |

