Search Count: 19
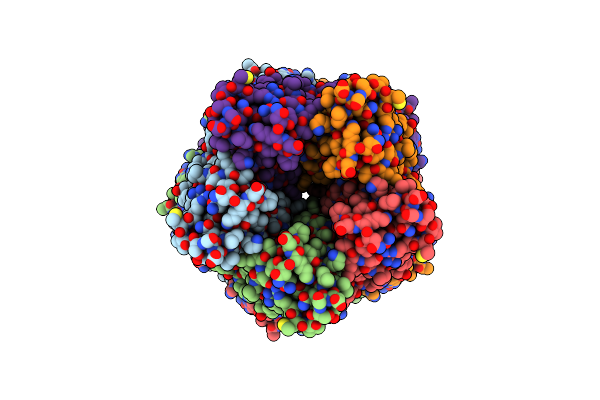 |
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
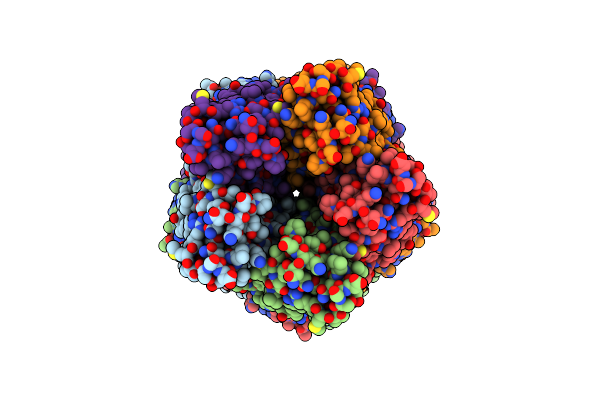 |
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
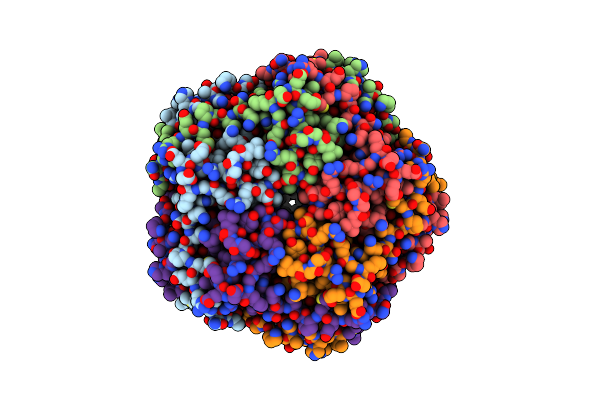 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
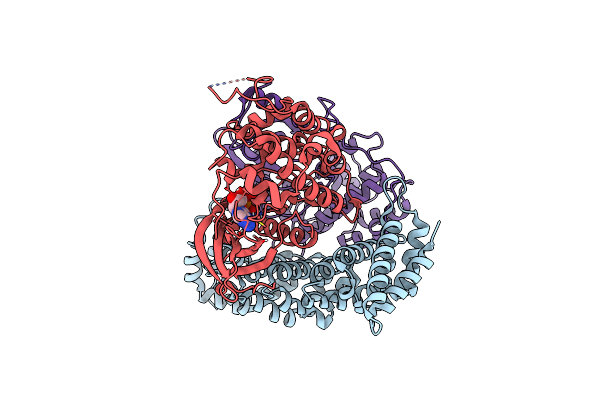 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-03-04 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Nectria haematococca (strain 77-13-4 / atcc mya-4622 / fgsc 9596 / mpvi)
Method: ELECTRON MICROSCOPY Release Date: 2019-11-06 Classification: LIPID TRANSPORT Ligands: CA |
 |
Organism: Escherichia coli (strain k12), Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
Organism: Escherichia coli (strain k12), Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
Organism: Escherichia coli (strain k12), Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
Organism: Escherichia coli (strain k12), Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
Crystal Structure Of The E148A Mutant Clc-Ec1 In The Presence Of 50Mm Bromoacetate
Organism: Escherichia coli (strain k12), Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: BXA |
 |
Crystal Structure Of The E148D/R147A/F317A Mutant In Presence Of 200 Mm Nabr
Organism: Escherichia coli ms 117-3, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: BR |
 |
Organism: Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: BR |
 |
Organism: Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: BR |
 |
Crystal Structure Of The E148D/R147A/F317A Mutant Clc-Ec1 In The Presence Of 20 Mm Nabr
Organism: Escherichia coli ms 117-3, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: BR |
 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: ELECTRON MICROSCOPY Release Date: 2019-02-06 Classification: LIPID TRANSPORT |
 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: ELECTRON MICROSCOPY Release Date: 2019-02-06 Classification: LIPID TRANSPORT Ligands: CA |
 |
Aftmem16 Reconstituted In Nanodiscs In The Presence Of Ca2+ And Ceramide 24:0
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: ELECTRON MICROSCOPY Release Date: 2019-02-06 Classification: LIPID TRANSPORT Ligands: CA, D12, D10, 8K6, HEX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-11-11 Classification: IMMUNE SYSTEM Ligands: CA, 57S, ACT, GOL |

