Search Count: 14
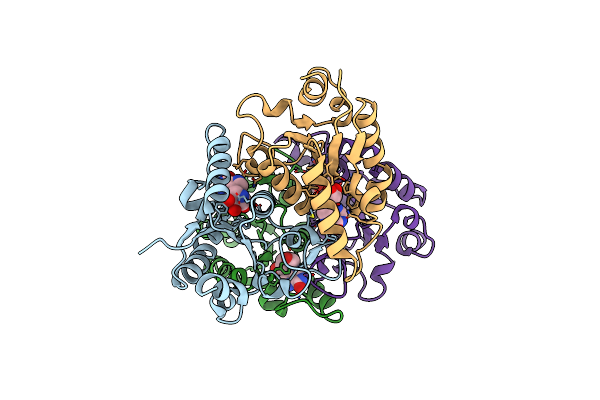 |
Nucleoside 2'Deoxyribosyltransferase From Chroococcidiopsis Thermalis Pcc 7203 Y7F Mutant Bound To Immh-Forodesine
Organism: Chroococcidiopsis thermalis pcc 7203
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: IMH |
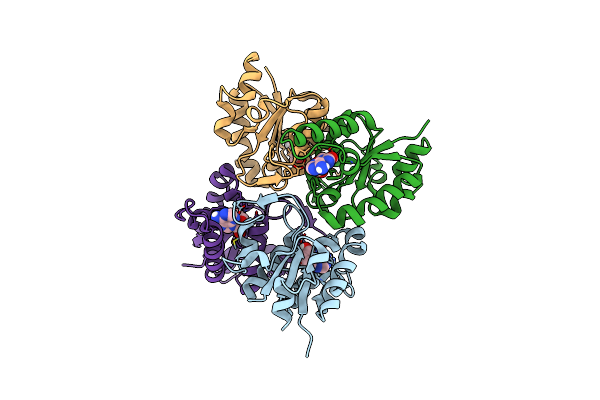 |
Nucleoside 2'Deoxyribosyltransferase From Chroococcidiopsis Thermalis Pcc 7203 Double Mutant Y7F A9S Bound To Cordycepin
Organism: Chroococcidiopsis thermalis pcc 7203
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 3AD |
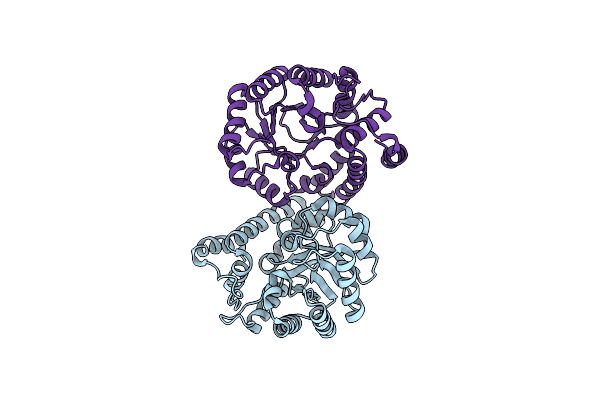 |
Crystal Structure Of The Wild-Type Staphylococcus Aureus N- Acetylneurminic Acid Lyase In Complex With Fluoropyruvate
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-11-18 Classification: LYASE |
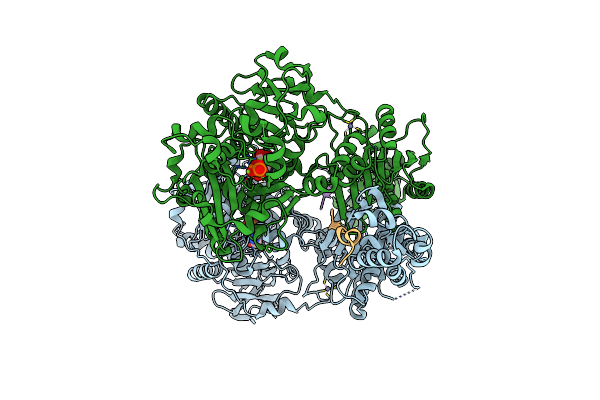 |
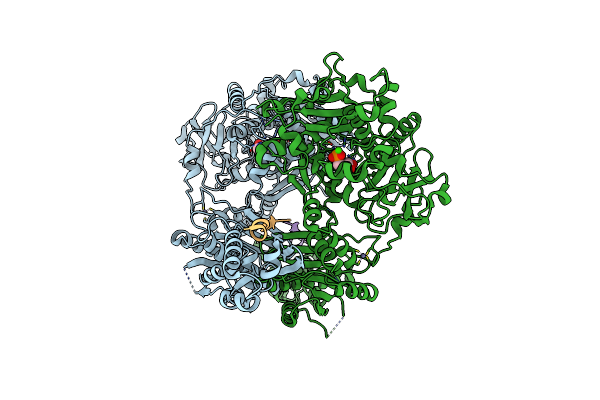
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4 |
 |
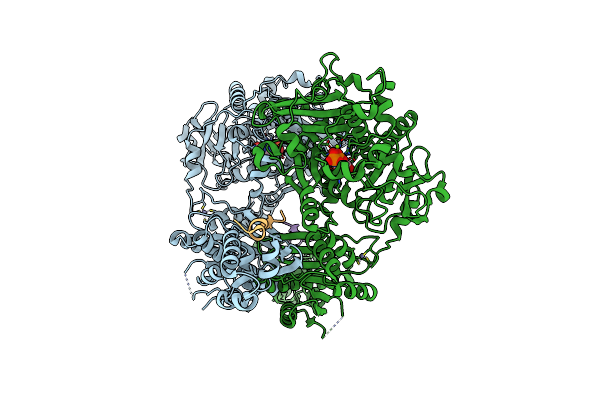
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, AMP, CA |
 |
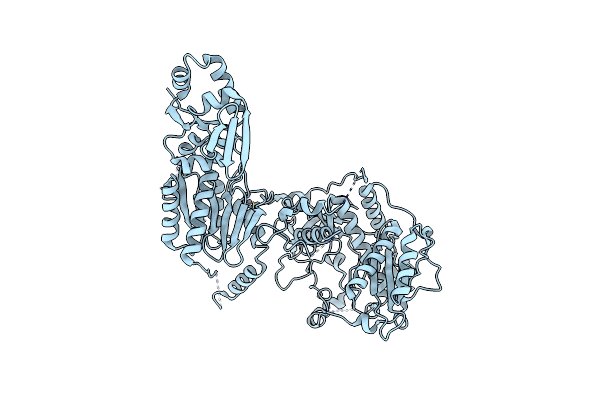
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, ANP, MG |
 |
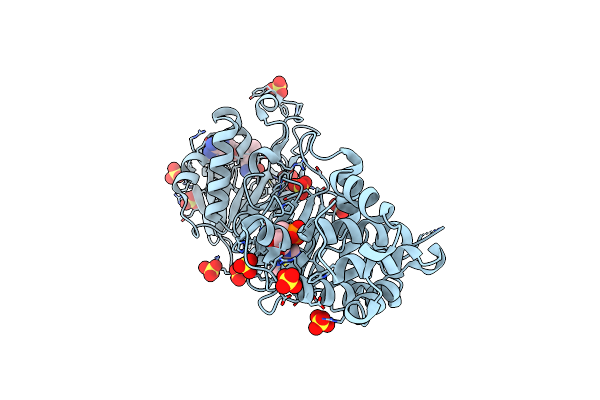
Organism: Uncultured prochloron sp. 06037a
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-11-06 Classification: ISOMERASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-09-26 Classification: TRANSFERASE Ligands: AMP, SAM, SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2012-09-26 Classification: TRANSFERASE Ligands: SAM, SO4, CL, LY2 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2012-09-26 Classification: TRANSFERASE Ligands: SAM, SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2012-09-26 Classification: TRANSFERASE Ligands: ATP, SAM, MG |
 |
Crystal Structures Of (S)-(-)-Blebbistatin Analogs Bound To Dictyostelium Discoideum Myosin Ii
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-02-19 Classification: CONTRACTILE PROTEIN Ligands: MG, VO4, BL4, ADP |
 |
Crystal Structures Of (S)-(-)-Blebbistatin Analogs Bound To Dictyostelium Discoideum Myosin Ii
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-02-19 Classification: CONTRACTILE PROTEIN Ligands: MG, VO4, ADP, EDO, BL6 |
 |
Crystal Structures Of (S)-(-)-Blebbistatin Analogs Bound To Dictyostelium Discoideum Myosin Ii
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-02-19 Classification: CONTRACTILE PROTEIN Ligands: MG, VO4, ADP, BL7, EDO |

