Search Count: 1,107
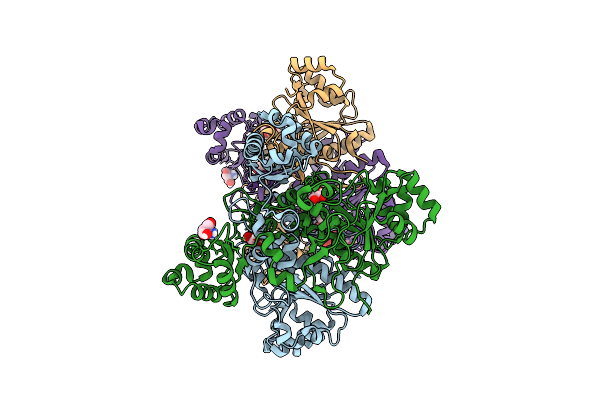 |
Organism: Aromatoleum aromaticum ebn1
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: GOL, TRS, COA |
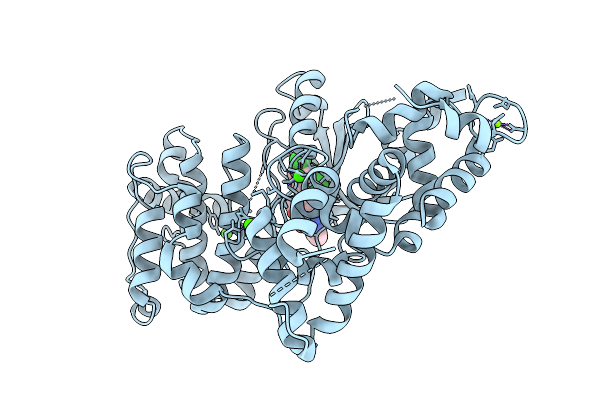 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Win-3-115
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: CA, A1BLB, MG, CL |
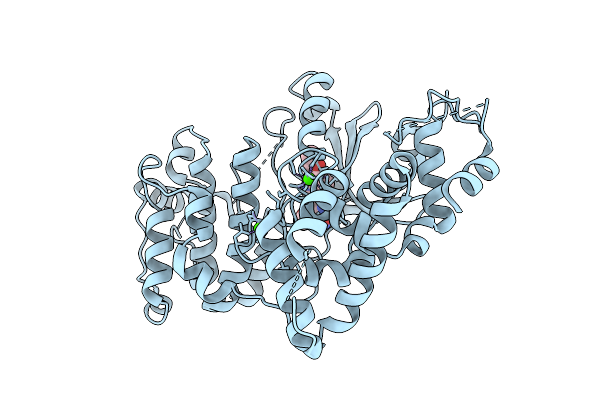 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Bki-1606
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1BLC, CA |
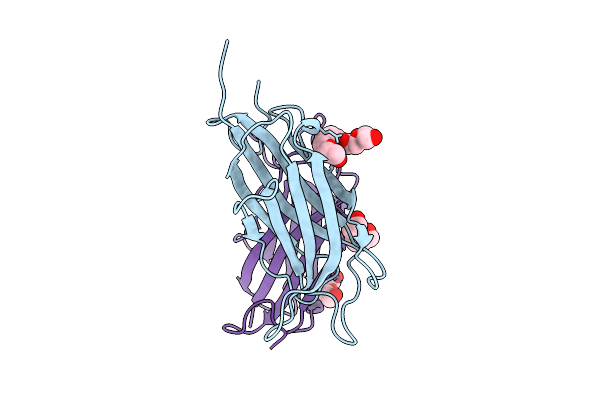 |
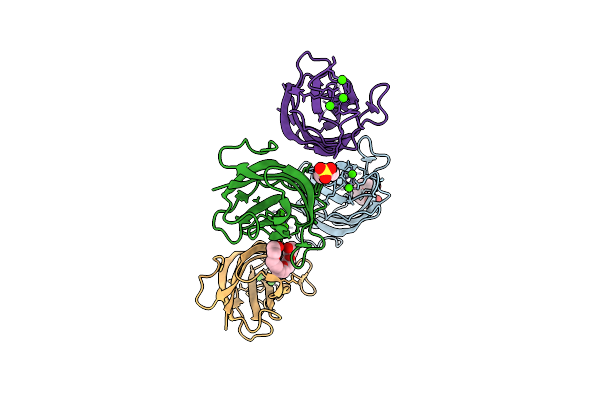
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: 1PE, CL, PEG, PGE |
 |
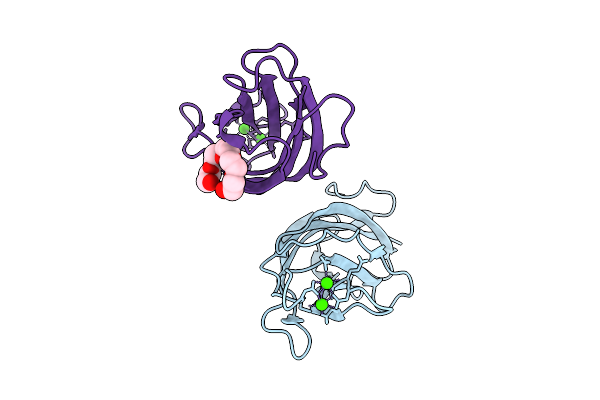
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PG4, CA, MES, 1PE |
 |
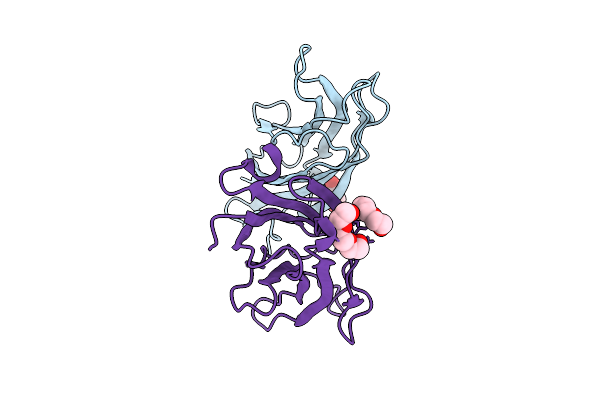
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (Orthorhombic P Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, 1PE |
 |
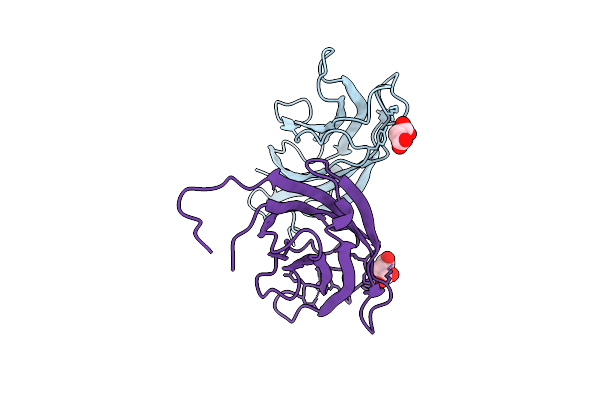
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: GOL, PGE |
 |
Crystal Structure Of A Malonate Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: MLI, CL, NA |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P61 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, CL, PG4 |
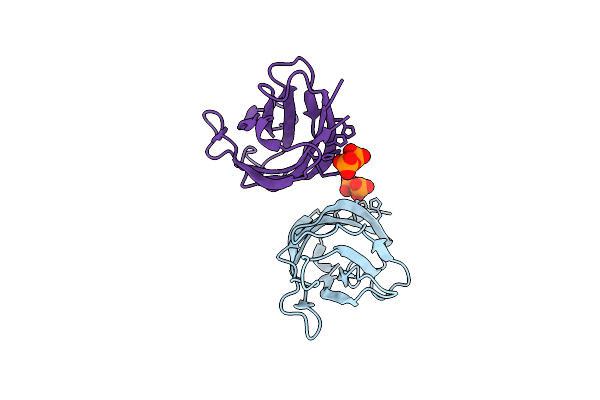 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis In Complex With Pyrophosphate
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PPV |
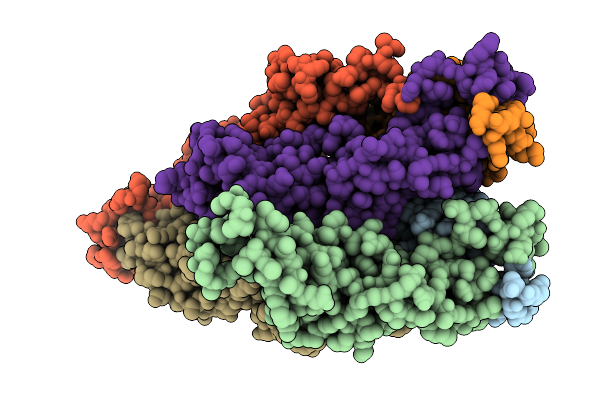 |
Organism: Brucella abortus 2308
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: STRUCTURAL PROTEIN Ligands: CL |
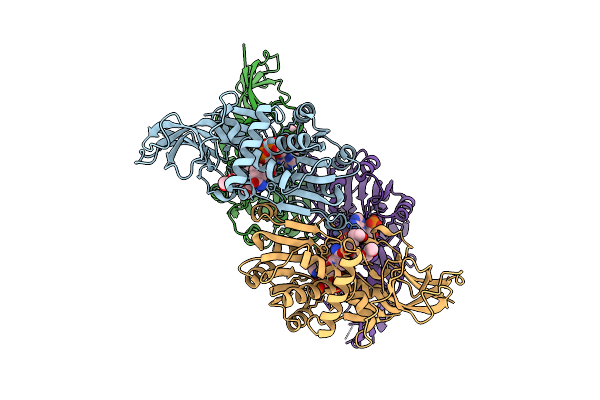 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: CL, NAP, PG6 |
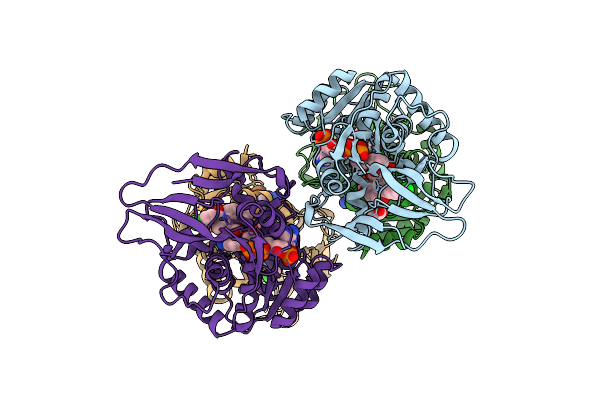 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp And Indomethacin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: ACT, IMN, NAP, GOL |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp And Indomethacin (Orthorhombic P Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: CL, SO4, GOL, IMN, NAP, PG4 |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad And Glyceraldehyde-3-Phosphate
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: G3H, PEG, NAD, NA, PGE, CL, GOL, EPE |
 |
Crystal Structure Of Formyl-Coenzyme A Transferase From Brucella Melitensis In Complex With Succinate
Organism: Brucella melitensis 16m1w
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: SIN |
 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis In Complex With 3-Phosphoglyceric Acid
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: CL, 3PG, PG4, SO4 |
 |
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae (Orthorhombic C Form)
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: CL |
 |
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae In Complex With Atp
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: SO4, CL, ATP |
 |
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae In Complex With O5'-(L-Glutamyl-Sulfamoyl)-Adenosine
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: GSU, CL |

