Search Count: 15
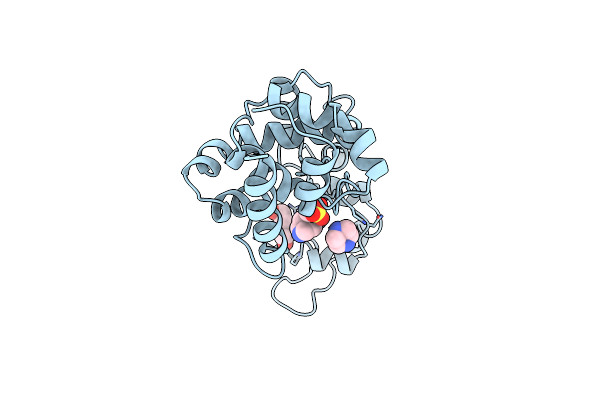 |
Crystal Structure Of Endolysin Gp46 From Pseudomonas Aeruginosa Bacteriophage Vb_Paem_Ktn6
Organism: Bacteriophage sp.
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: EPE, PEG |
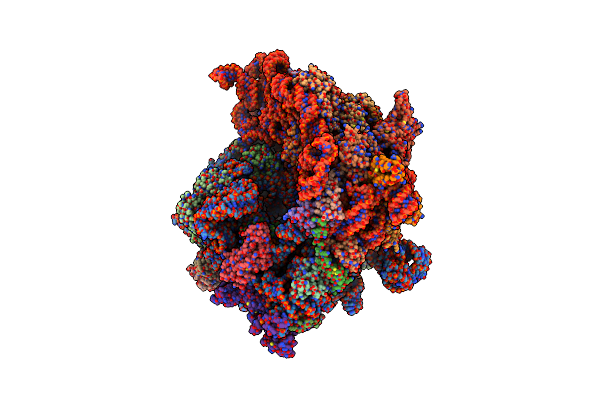 |
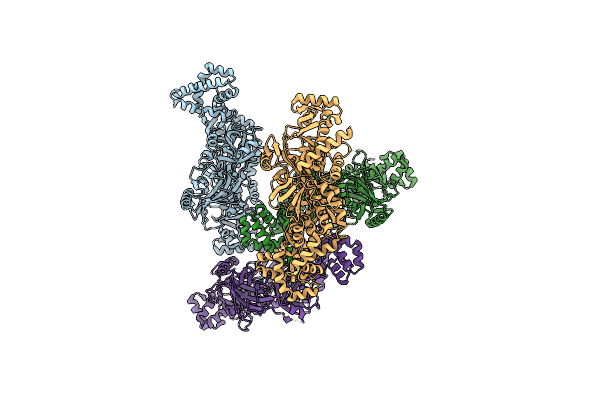
Organism: Pseudomonas aeruginosa pao1, Pseudomonas phage phikz
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: RIBOSOME Ligands: MG |
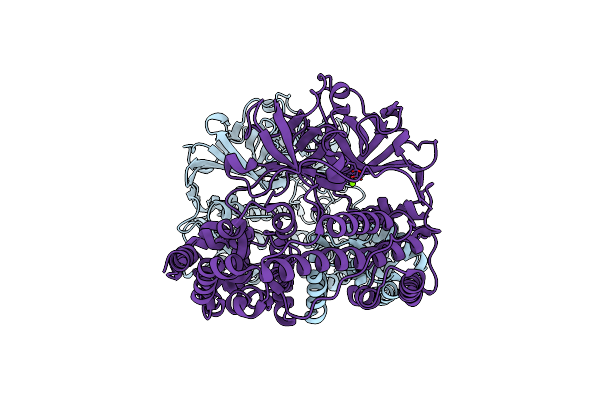 |
Organism: Thermus virus p74-26
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-01-17 Classification: REPLICATION Ligands: MG |
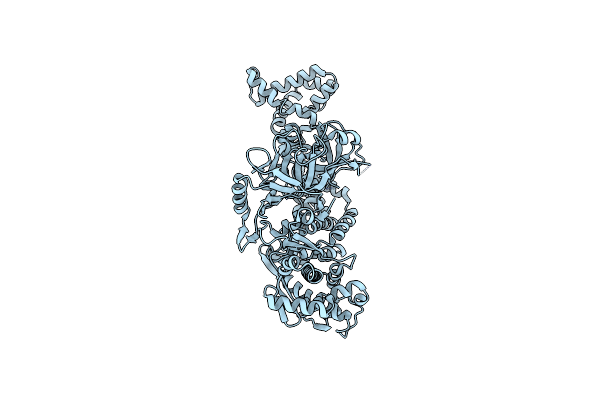 |
Organism: Oshimavirus p2345
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2023-10-11 Classification: TRANSCRIPTION |
 |
Organism: Thermus virus p23-45
Method: X-RAY DIFFRACTION Resolution:4.41 Å Release Date: 2023-10-11 Classification: TRANSCRIPTION Ligands: MG |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadph And Kalimantacin A (Batumin)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-04-01 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, KAL |
 |
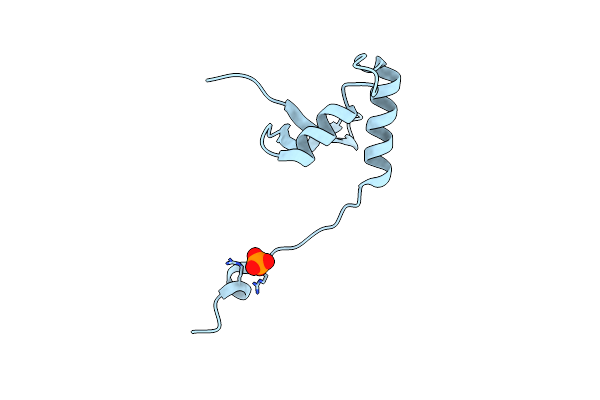
Crystal Structure Of S. Aureus Fabi In Complex With Nadph And Kalimantacin B
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-04-01 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, N5H |
 |
Organism: Pseudomonas phage luz7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-30 Classification: VIRAL PROTEIN Ligands: PO4 |
 |
Organism: Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-08-03 Classification: HYDROLASE INHIBITOR Ligands: ARG, K |
 |
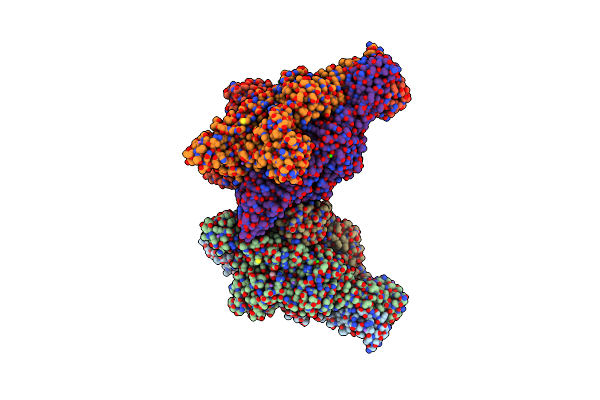
Crystal Structure Of Gp37(Dip) From Bacteriophage Phikz Bound To Rnase E Of Pseudomonas Aeruginosa
Organism: Pseudomonas phage phikz, Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-08-03 Classification: HYDROLASE/INHIBITOR Ligands: HOH |
 |
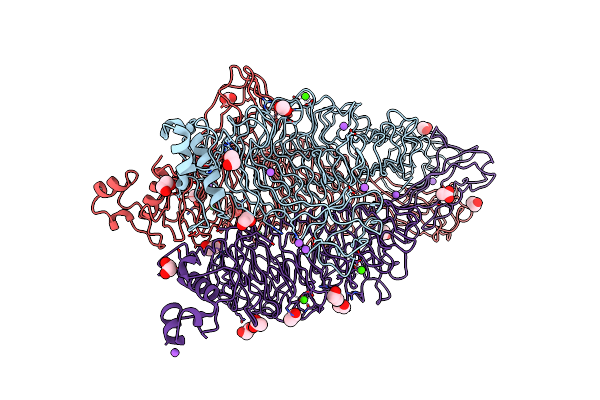
Crystal Structure Of The Tailspike Protein Gp49 From Pseudomonas Phage Lka1
Organism: Pseudomonas phage lka1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-11-18 Classification: STRUCTURAL PROTEIN Ligands: EDO, NA, CA |
 |
Organism: Pseudomonas phage phi297
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-11-18 Classification: LYASE Ligands: CA, NA, EDO, ACT |
 |
Organism: Pseudomonas fluorescens
Method: SOLUTION NMR Release Date: 2012-02-15 Classification: BIOSYNTHETIC PROTEIN |
 |
Elucidation Of The Substrate Specificity And Protein Structure Of Abfb, A Family 51 Alpha-L-Arabinofuranosidase From Bifidobacterium Longum.
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-05-19 Classification: HYDROLASE |

