Search Count: 22
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Bace-1 In Complex With Active Site Inhibitor Grl-8234 And Exosite Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: BSD |
 |
Crystal Structure Of Bace-1 In Complex With Active Site And Exosite Binding Peptide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-07-12 Classification: RIBOSOMAL PROTEIN |
 |
Structure Of The Viral Immunoevasin M12 (Smith) Bound To The Natural Killer Cell Receptor Nkr-P1B (B6)
Organism: Mus musculus, Murid herpesvirus 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-05-24 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: IOD |
 |
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm)
Organism: Pseudomonas mendocina (strain ymp)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, ETX, NA |
 |
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm) In Complex With The Dimethylamine Substrate
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, DMN, PEG, NA |
 |
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm) In Complex With The Dimethylamine Substrate
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, DMN, NO, PEG, NA |
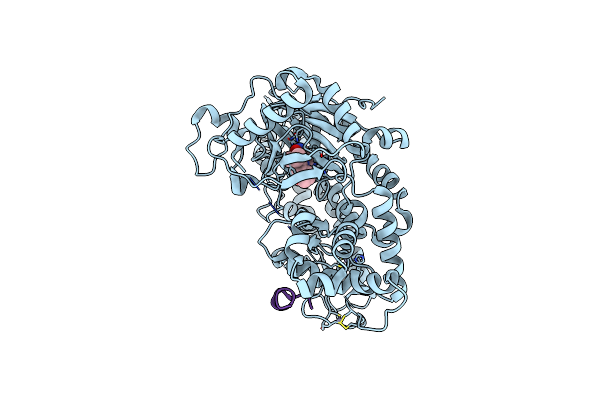 |
Structure Of Histone Lysine Demethylase Kdm5A In Complex With Selective Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2016-05-18 Classification: Oxidoreductase/Inhibitor Ligands: NI, ZN, 50P |
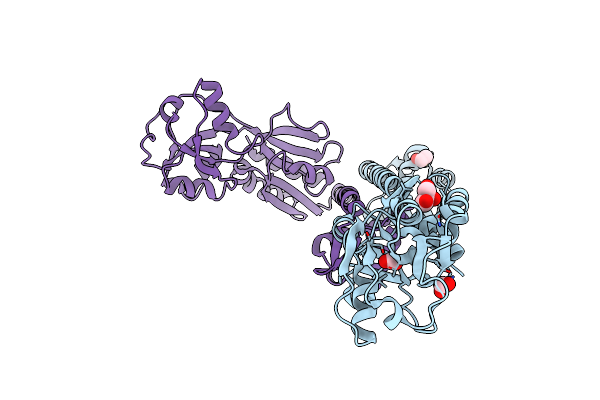 |
Organism: Comamonas testosteroni
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-01-26 Classification: Transcription regulator Ligands: BME, GOL, FMT, CL |
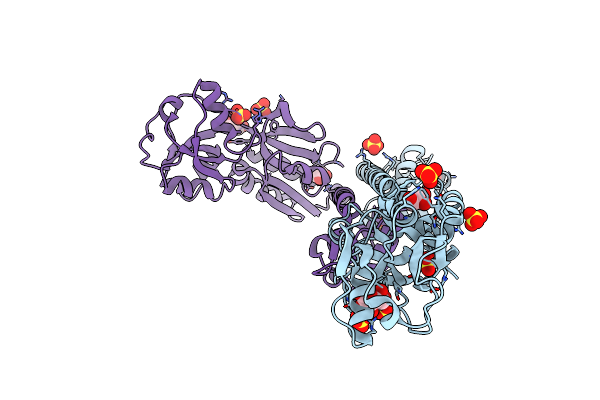 |
Organism: Comamonas testosteroni
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-01-26 Classification: Transcription regulator Ligands: SO4, GOL, ASC |
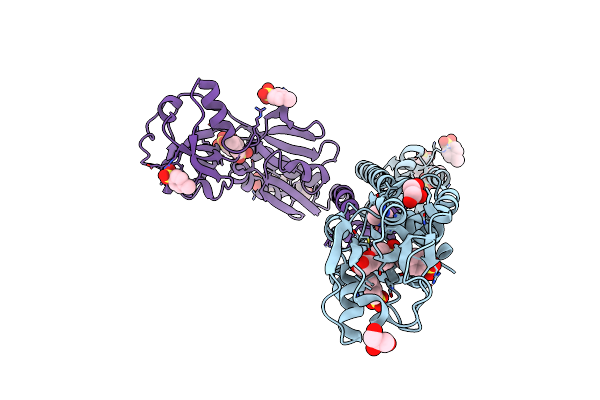 |
Organism: Comamonas testosteroni
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-01-26 Classification: Transcription regulator Ligands: TSU, IMD, GOL, CL, FMT |
 |
Organism: Comamonas testosteroni
Method: X-RAY DIFFRACTION Resolution:7.10 Å Release Date: 2010-01-26 Classification: Transcription regulator |
 |
Crystal Structure Of D552A Dimethylglycine Oxidase Mutant Of Arthrobacter Globiformis In Complex With Tetrahydrofolate
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-14 Classification: OXIDOREDUCTASE Ligands: THG, MG, FAD |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2009-03-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-03-03 Classification: PROTEIN BINDING Ligands: MPD, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-03-03 Classification: PROTEIN BINDING Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2009-03-03 Classification: PROTEIN BINDING Ligands: PO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-03-03 Classification: PROTEIN BINDING |
 |

