Search Count: 23
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: A1AB5 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: GLU, A1AB5 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-03-02 Classification: IMMUNE SYSTEM |
 |
Organism: Xenopus laevis, Rattus norvegicus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.55 Å Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 Active Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 Non-Active2 Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 In Non-Active1 Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 In Non-Active2-Like Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: PROTEIN FIBRIL |
 |
Organism: Mnemiopsis leidyi
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: GLY, MG, SO4 |
 |
Organism: Mnemiopsis leidyi
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: GLY, MG, SO4 |
 |
Crystal Structure Of A Tetramer Of Glua2 Tr Mutant Ligand Binding Domains Bound With Glutamate At 1.26 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2016-01-13 Classification: TRANSPORT PROTEIN Ligands: GLU, PO4, PEG |
 |
Crystal Structure Of A Tetramer Of Glua2 Ligand Binding Domains Bound With Glutamate At 1.45 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-01-13 Classification: TRANSPORT PROTEIN Ligands: GLU, PO4, PG4, PEG |
 |
Glua2-L483Y-A665C Ligand-Binding Domain In Complex With The Antagonist Dnqx
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-14 Classification: TRANSPORT PROTEIN Ligands: DNQ, SO4 |
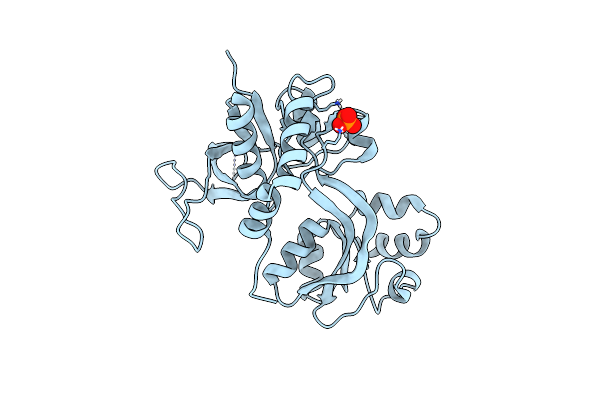 |
Crystal Structure Of The Nmda Receptor Glun1 Ligand Binding Domain Apo State
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2013-07-31 Classification: MEMBRANE PROTEIN Ligands: PO4 |
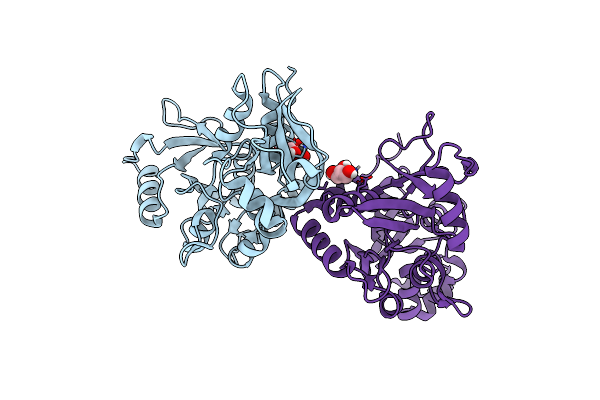 |
Crystal Structure Of The Nmda Receptor Glun3A Ligand Binding Domain Apo State
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2013-07-31 Classification: MEMBRANE PROTEIN Ligands: GOL |
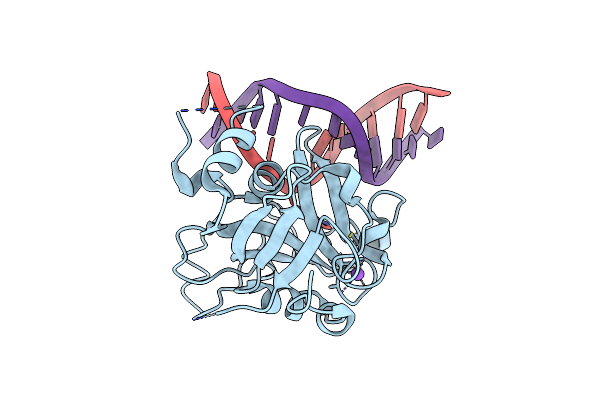 |
Crystal Structure Of The Human Aag Dna Repair Glycosylase Complexed With 1,N6-Ethenoadenine-Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-11 Classification: hydrolase/DNA |

