Search Count: 41
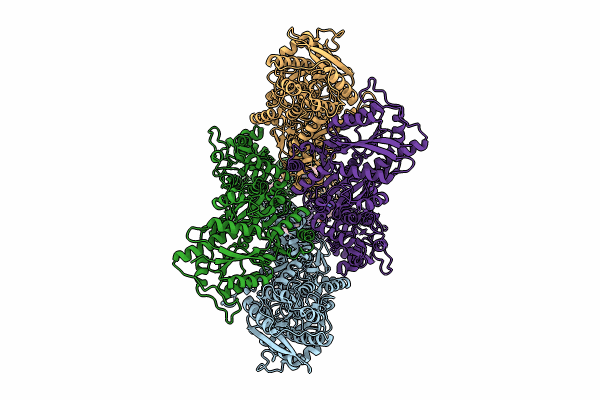 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: A1AB5 |
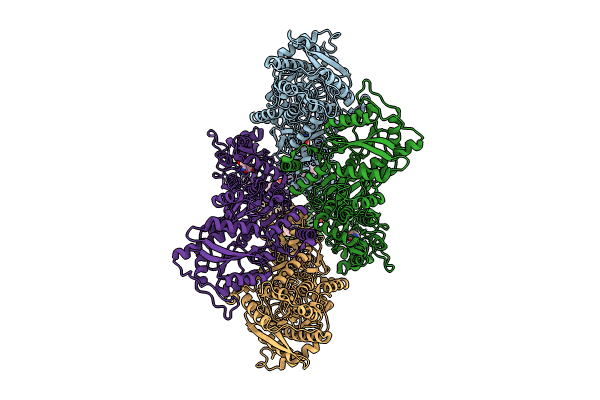 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: GLU, A1AB5 |
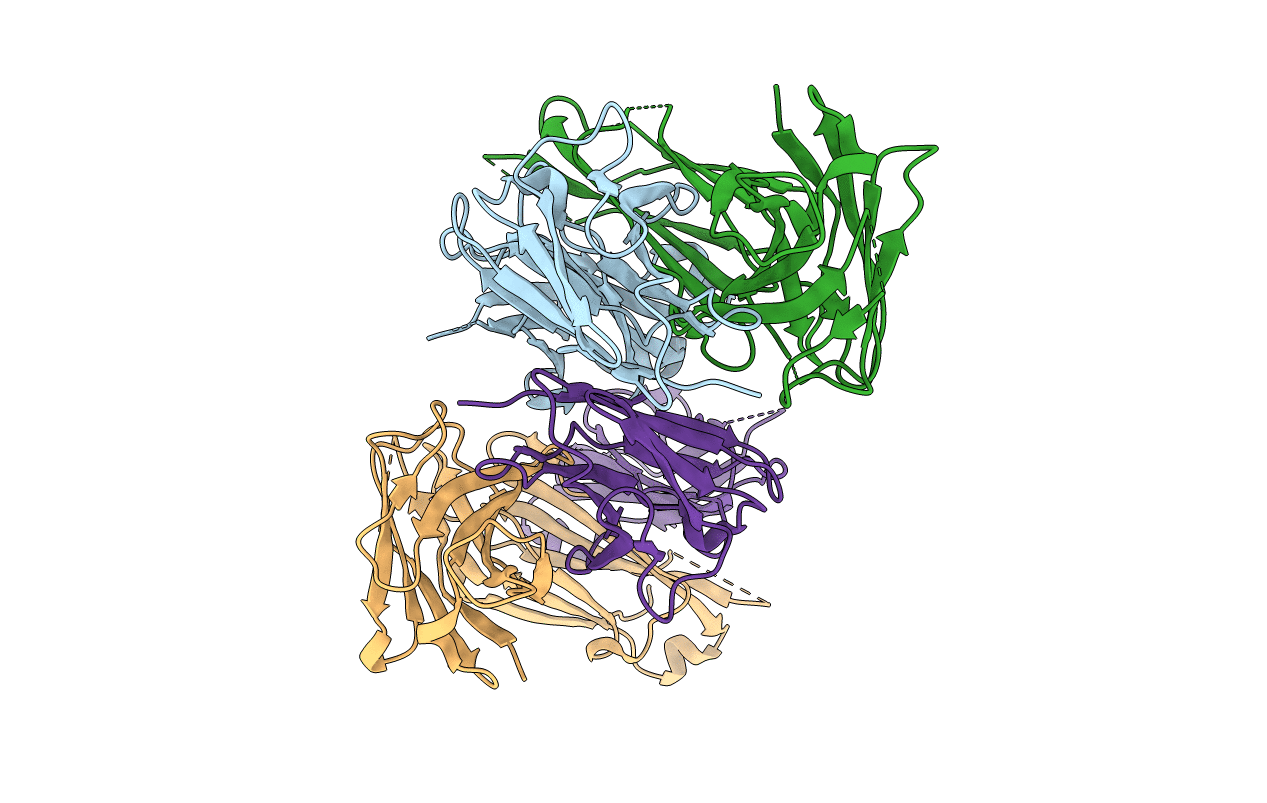 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-03-02 Classification: IMMUNE SYSTEM |
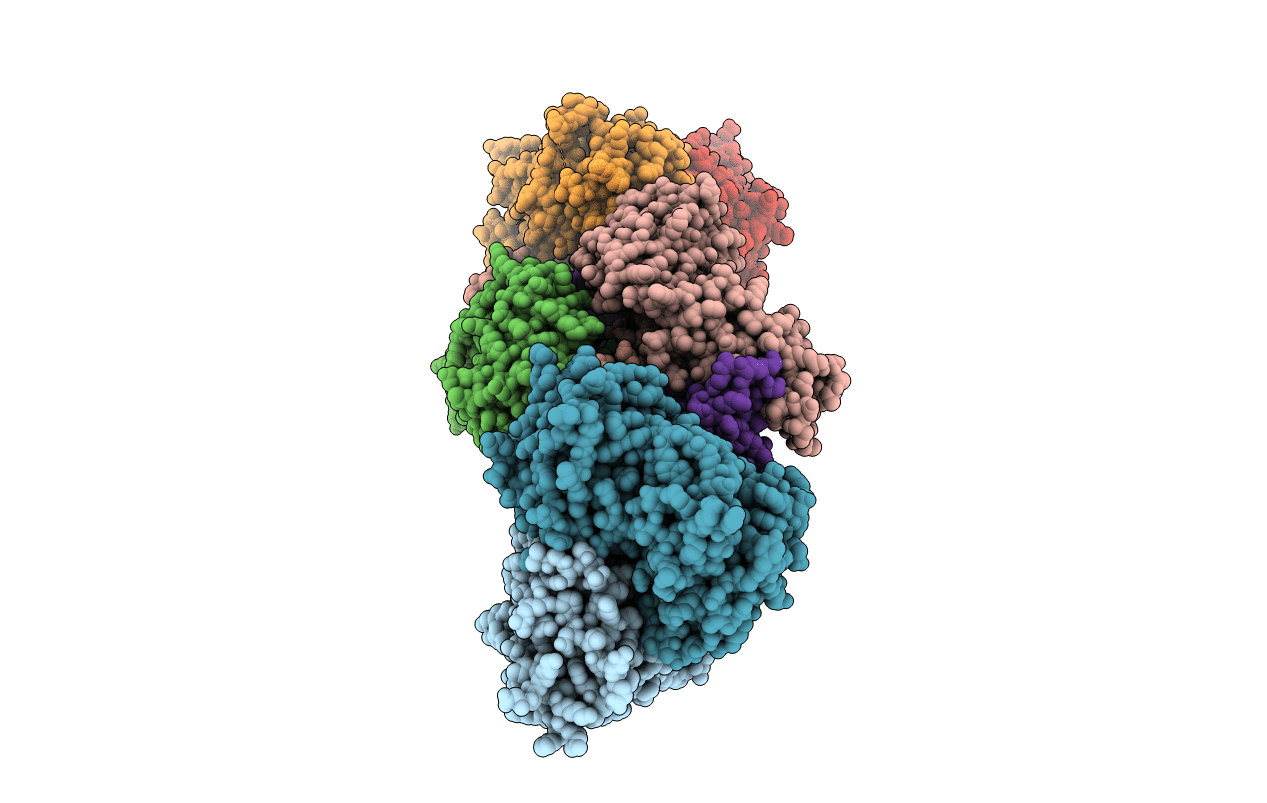 |
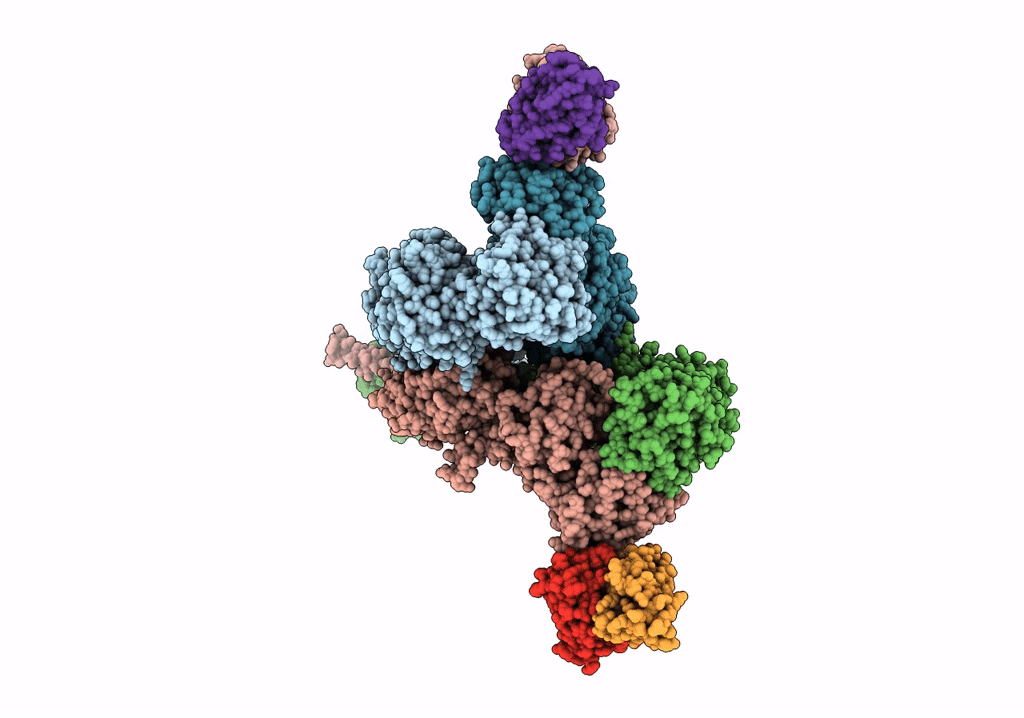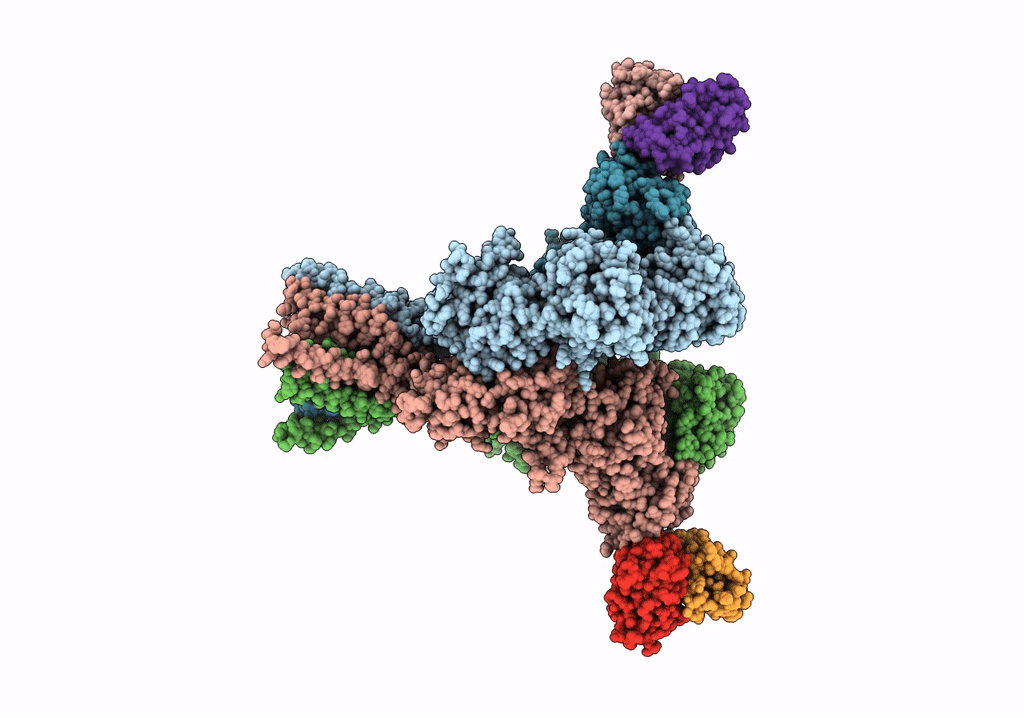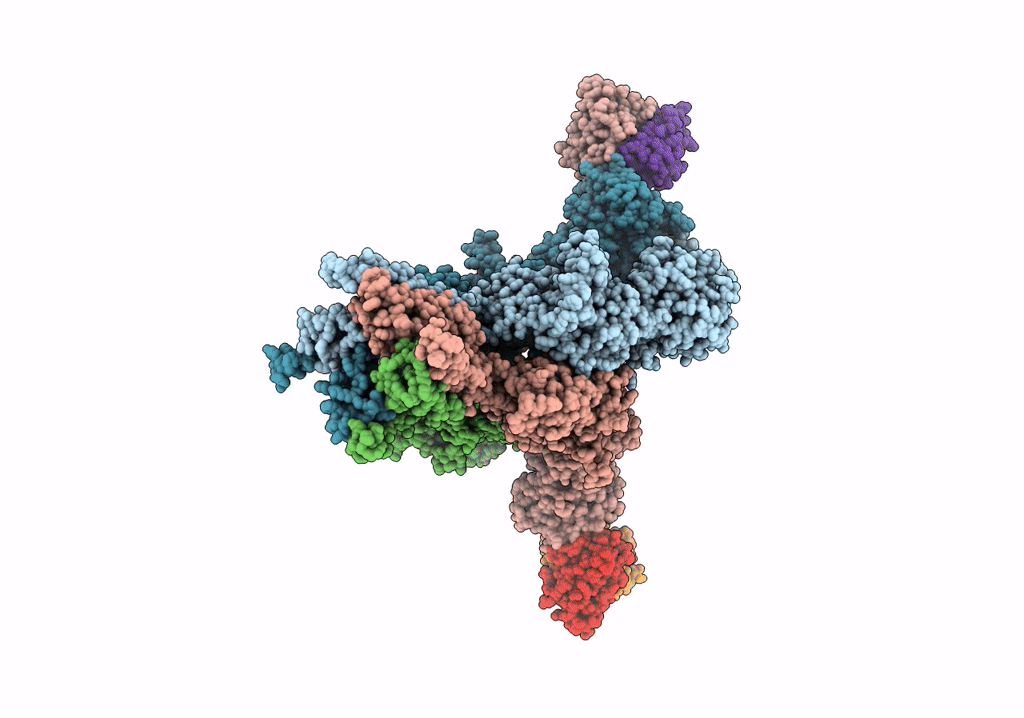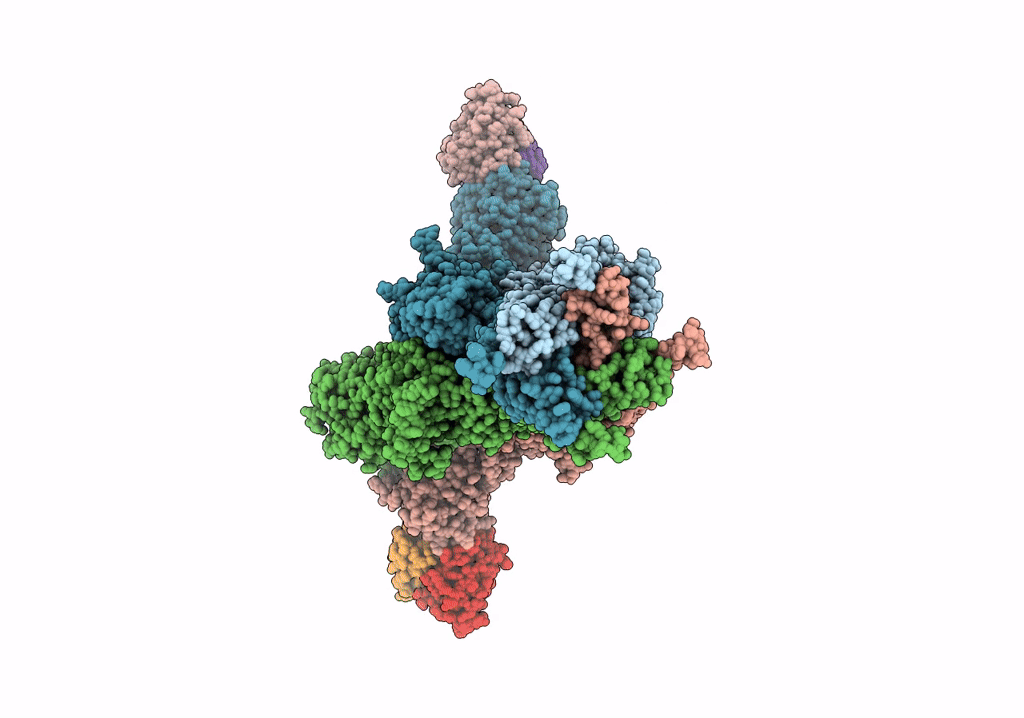
Organism: Xenopus laevis, Rattus norvegicus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.55 Å Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
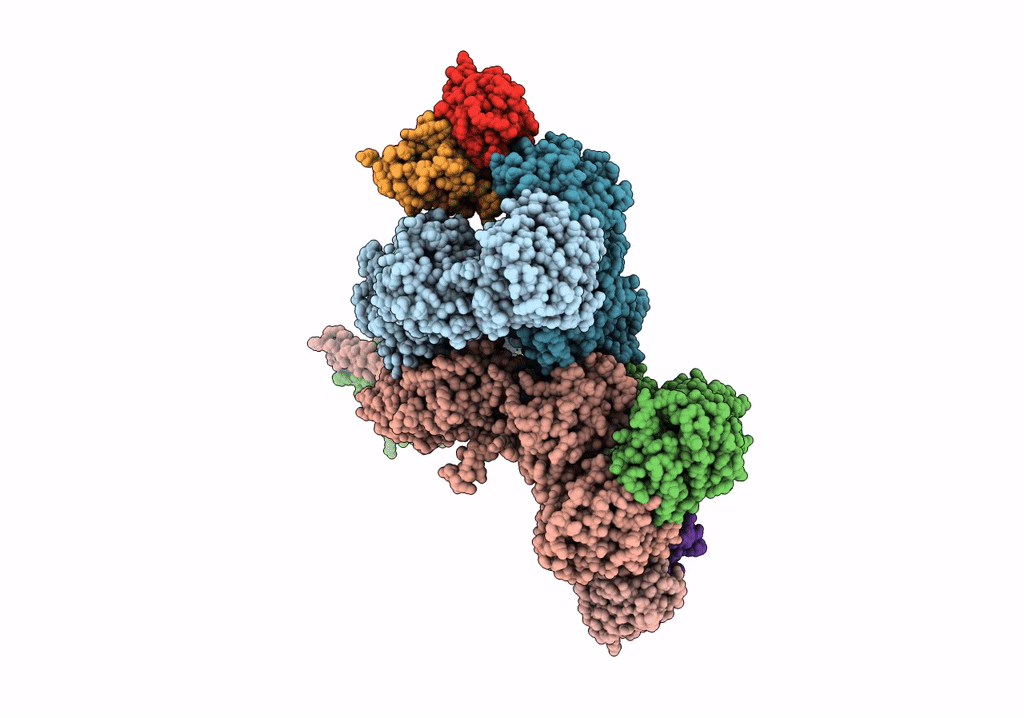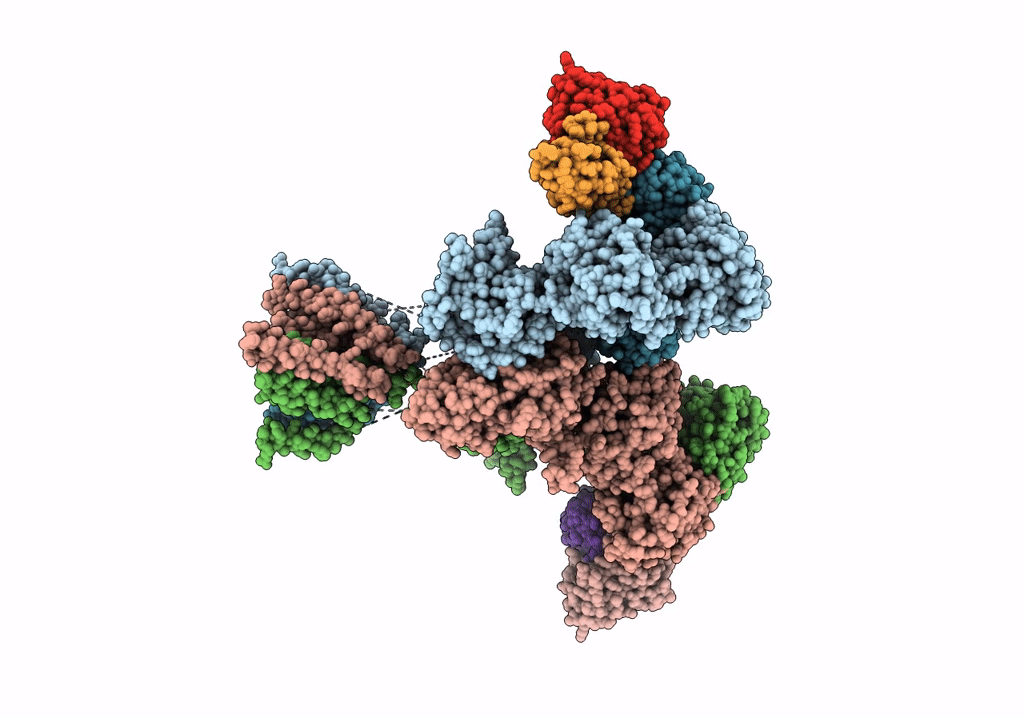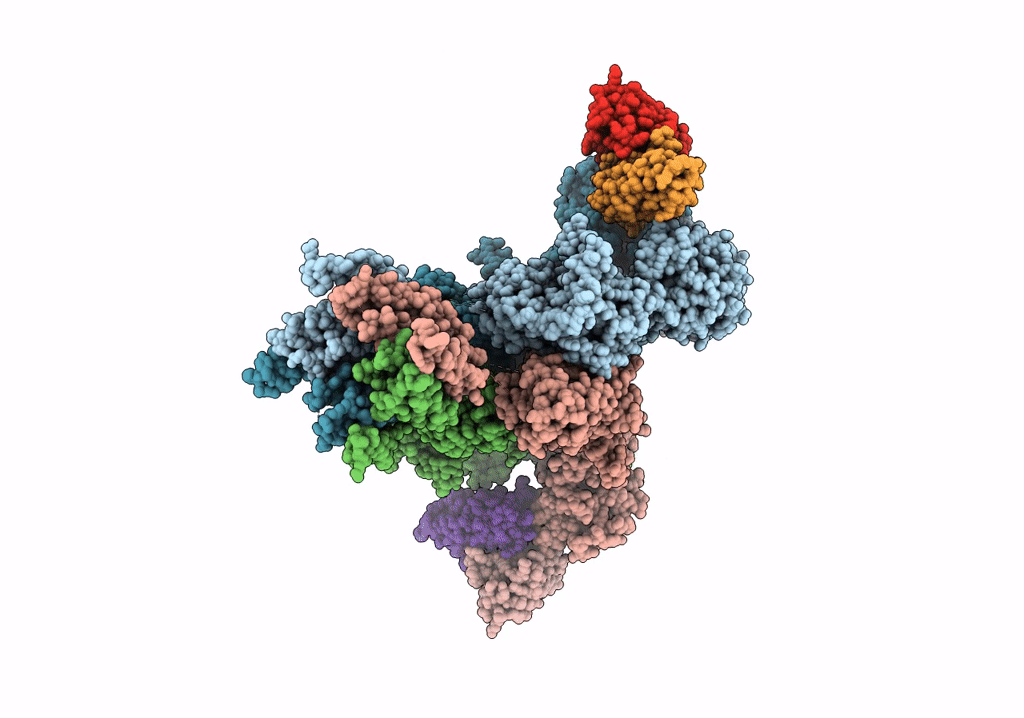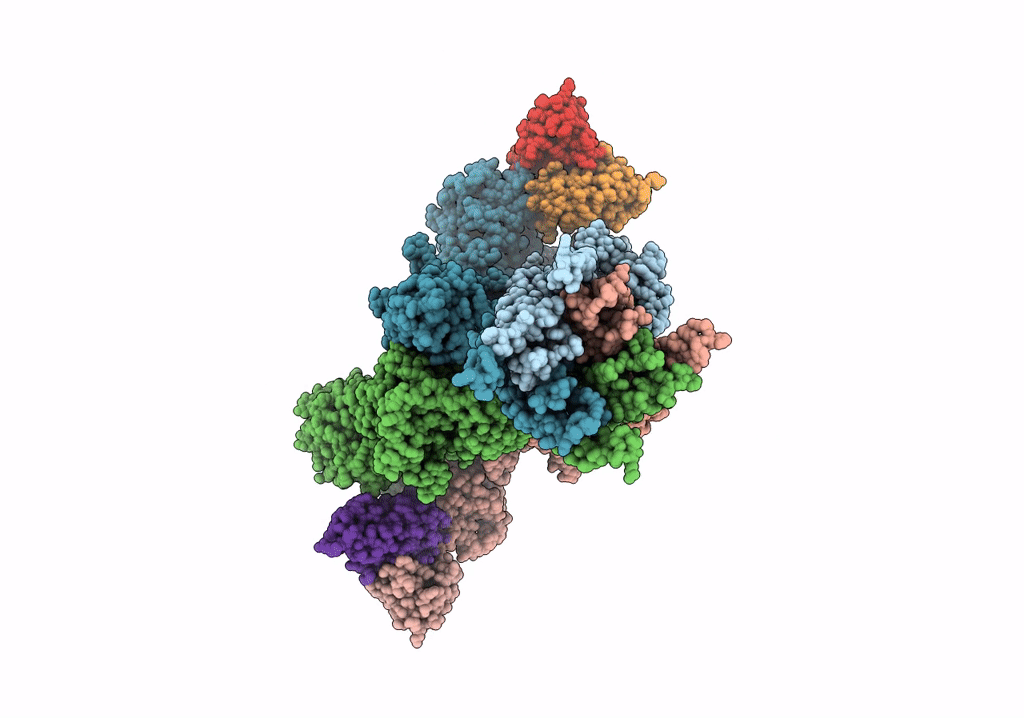 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 Active Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
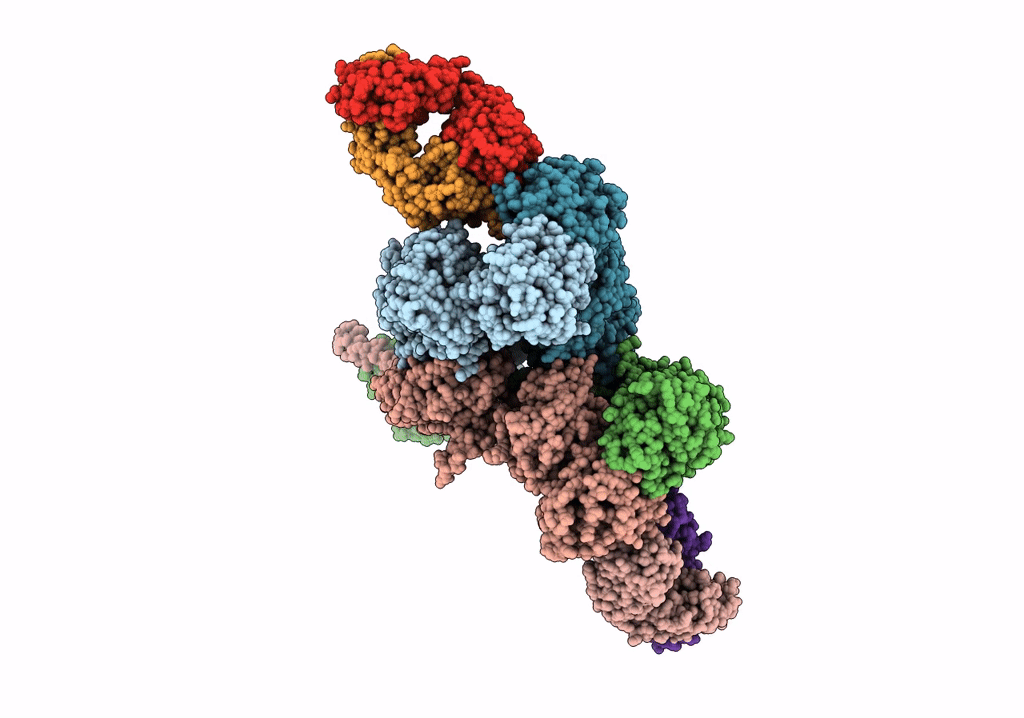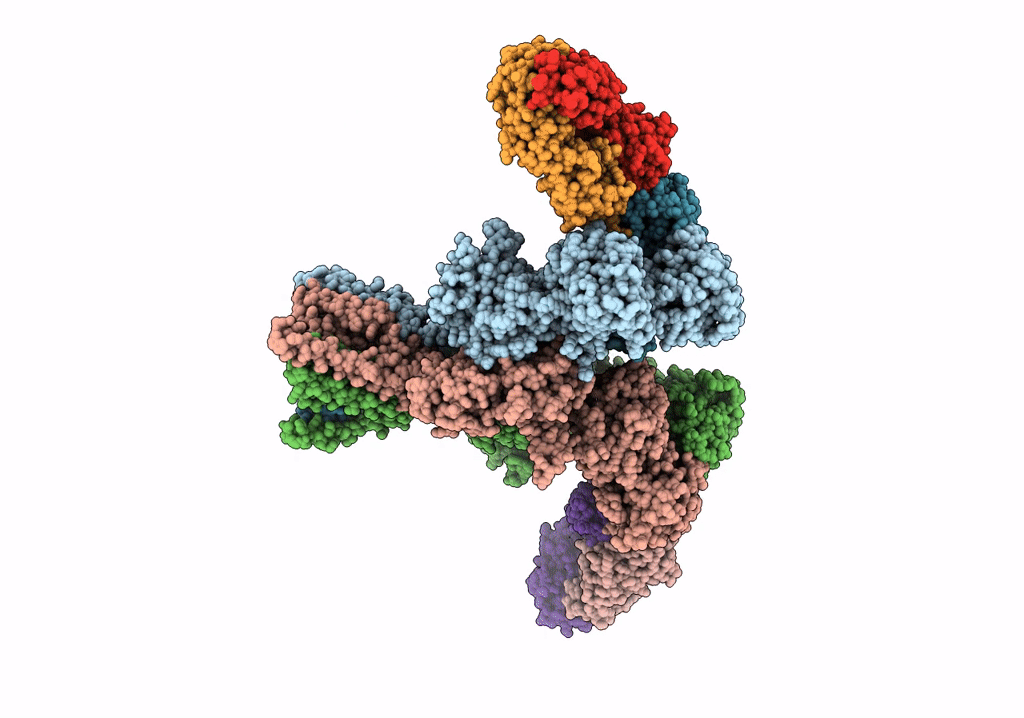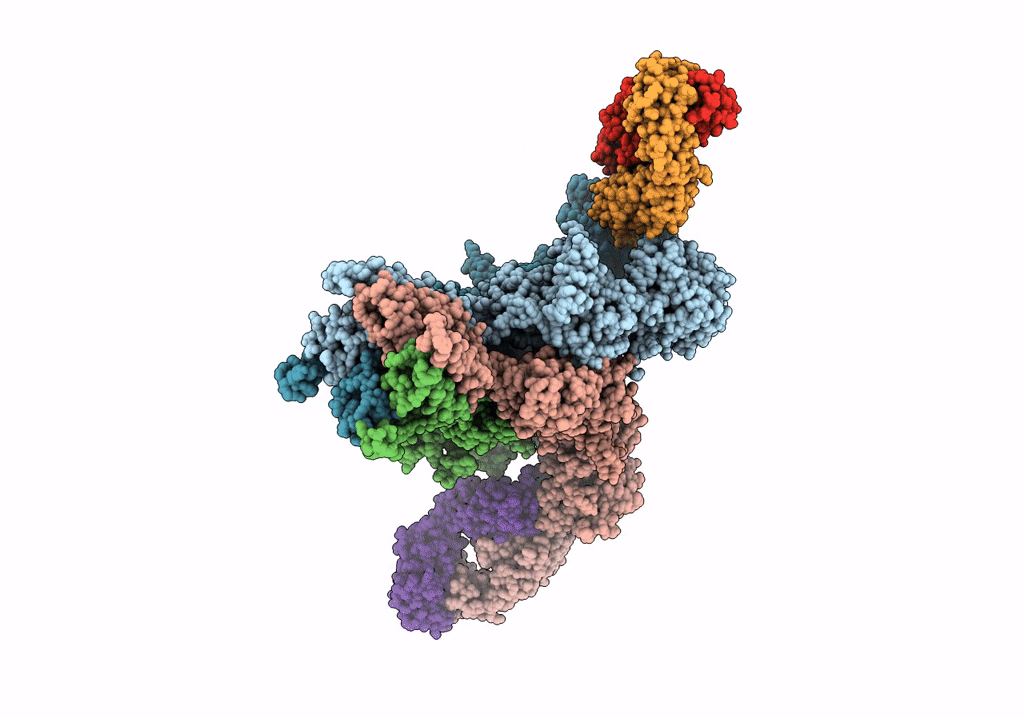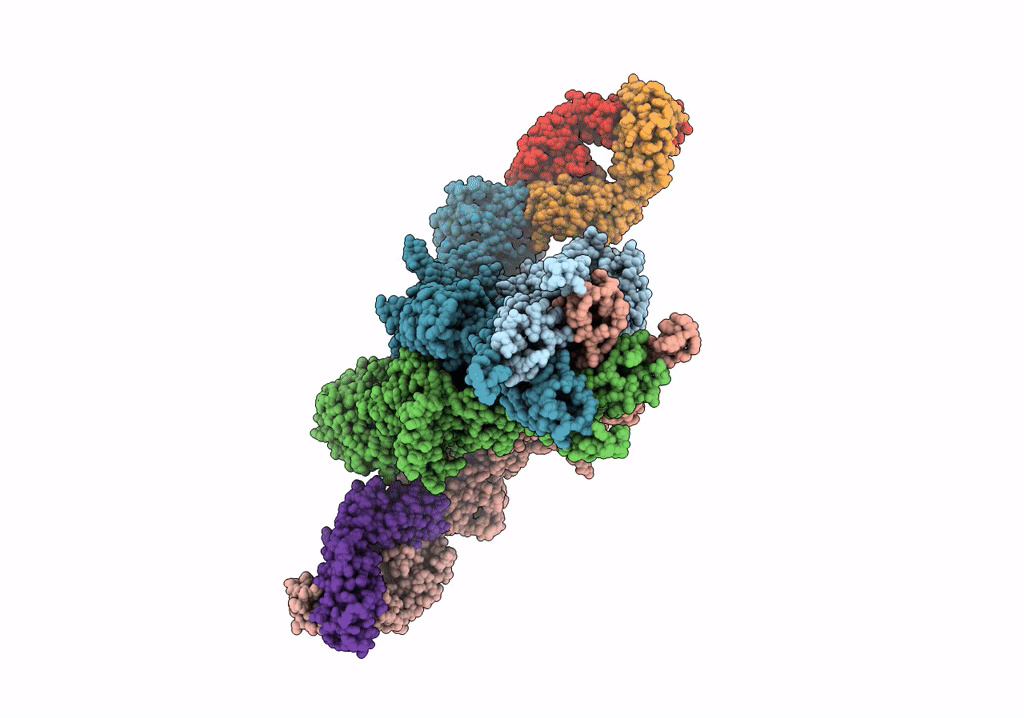
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 Non-Active2 Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 In Non-Active1 Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Glun1B-2B Nmdar In Complex With Fab5 In Non-Active2-Like Conformation
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN Ligands: SO4, 11D, AZI |
 |
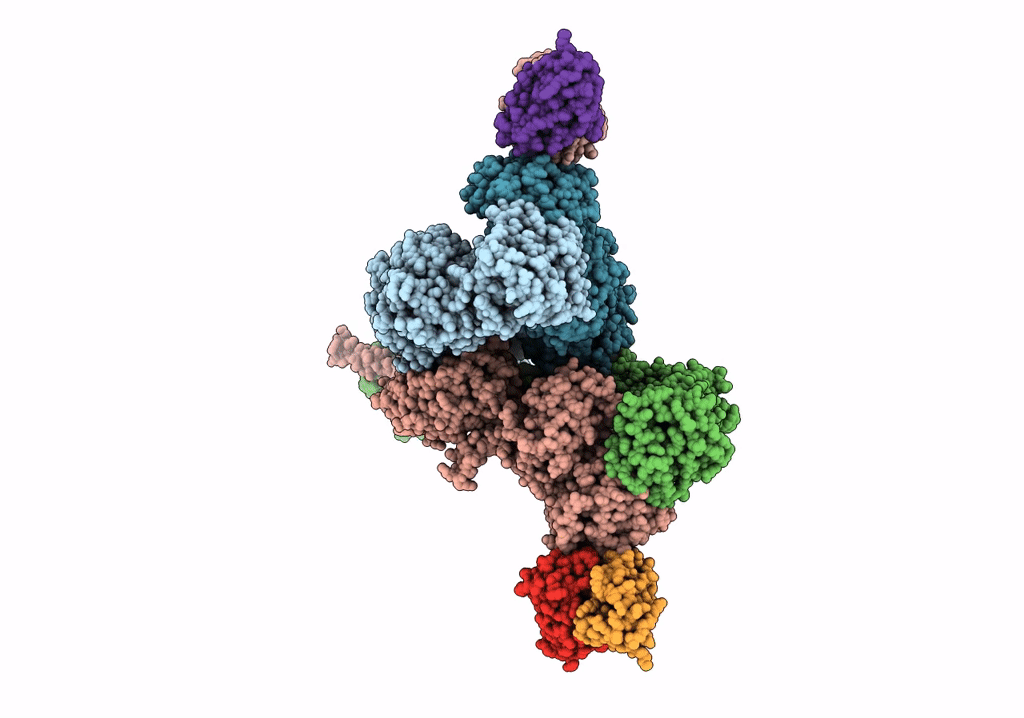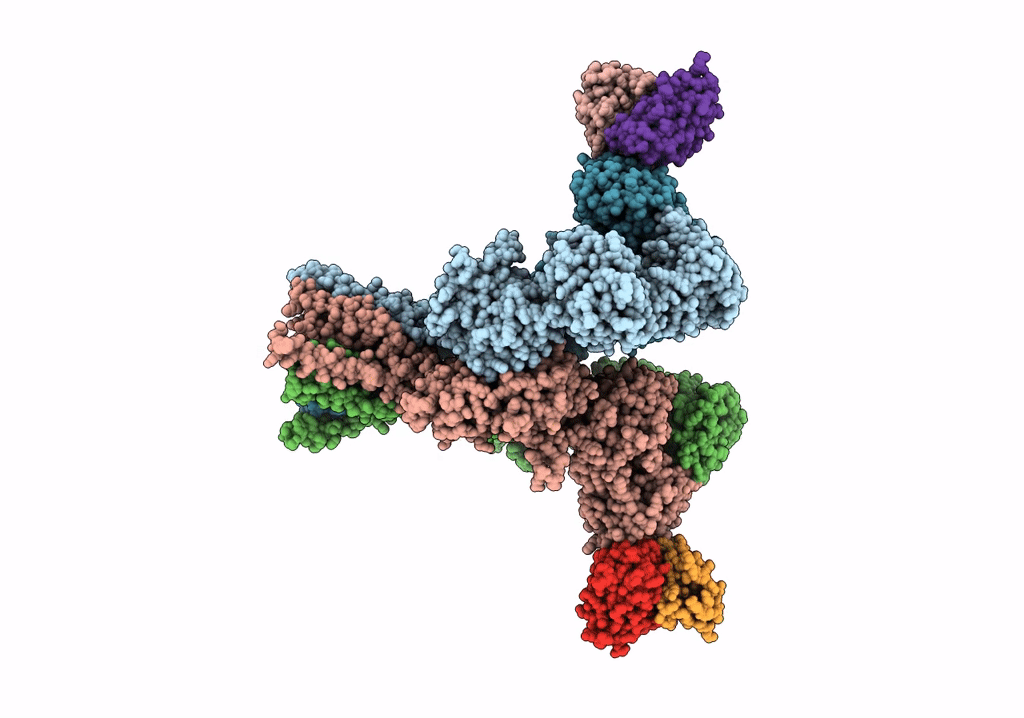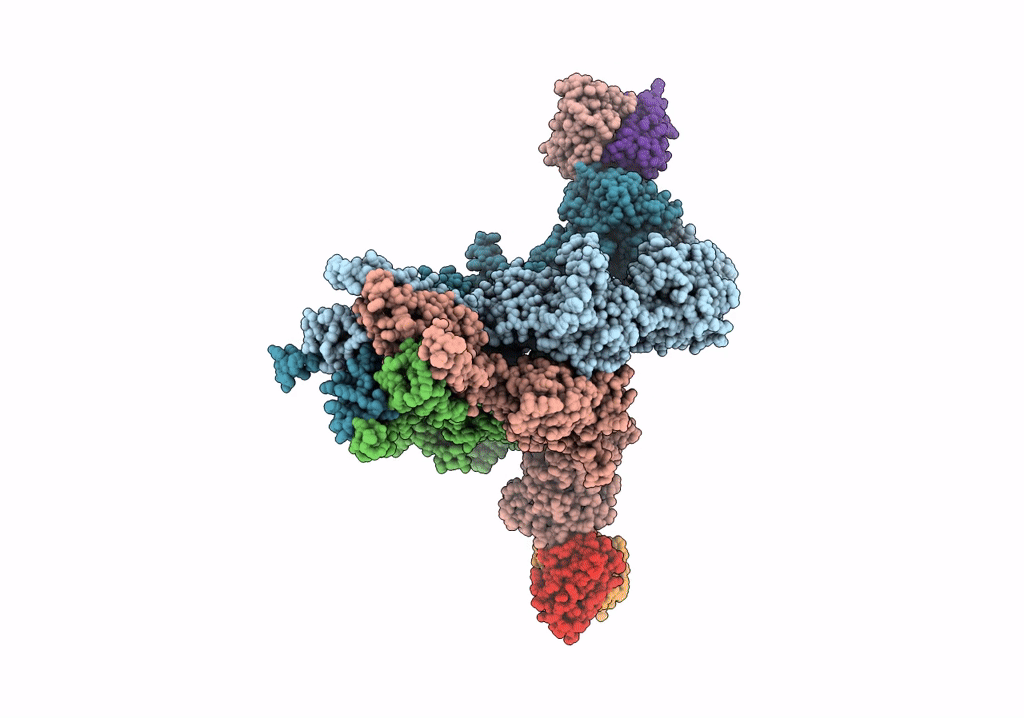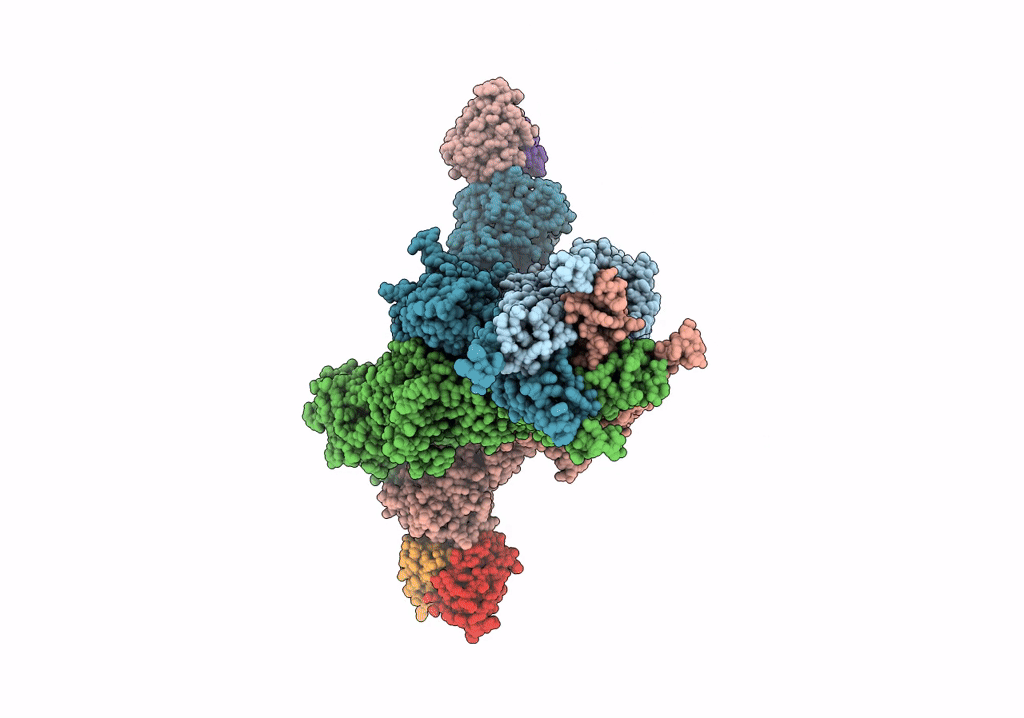
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
|
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
|
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-08-03 Classification: TRANSFERASE Ligands: 3QH, SO4 |
 |
Organism: Mnemiopsis leidyi
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: GLY, MG, SO4 |

