Search Count: 12
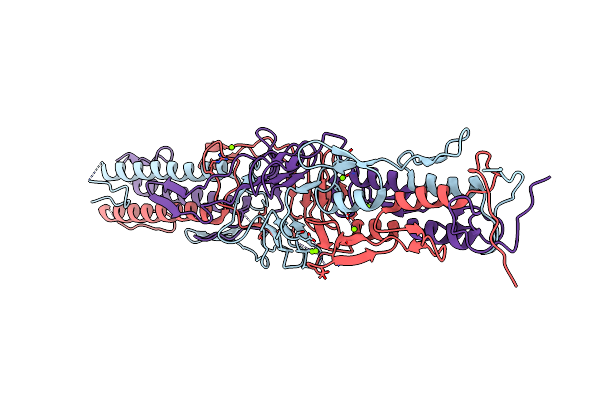 |
The Structure Of The C-Terminal Yada-Like Domain Of Bpsl2063 From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-07-22 Classification: TRANSPORT PROTEIN Ligands: MG |
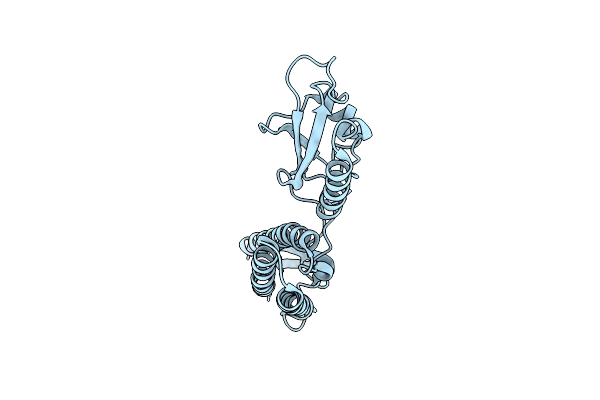 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-12-10 Classification: STRUCTURAL PROTEIN |
 |
Structural Characterization Using Sulfur-Sad Of The Cytoplasmic Domain Of Burkholderia Pseudomallei Pilo2Bp, An Actin-Like Protein Component Of A Type Ivb R64-Derivative Pilus Machinery.
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-04-23 Classification: MOTOR PROTEIN Ligands: PO4, K |
 |
Structural Characterization Using Sulfur-Sad Of The Cytoplasmic Domain Of Burkholderia Pseudomallei Pilo2Bp, An Actin-Like Protein Component Of A Type Ivb R64-Derivative Pilus Machinery.
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-04-23 Classification: MOTOR PROTEIN Ligands: K |
 |
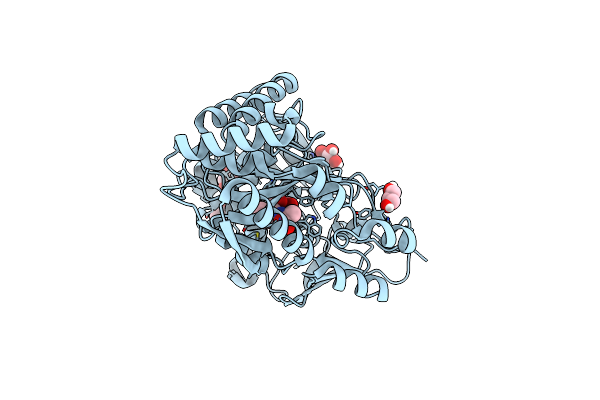
Crystal Structure Of The Peptidoglycan-Associated Lipoprotein From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-14 Classification: LIPID TRANSPORT Ligands: ACT |
 |
Organism: Burkholderia pseudomallei, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-07-04 Classification: PEPTIDE BINDING PROTEIN Ligands: GOL, CL |
 |
Crystal Structure Of The Mono-Zinc Metallo-Beta-Lactamase Vim-4 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, FLC, CIT |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, IFS, SO4, GOL |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, GOL, SO4, SDF |
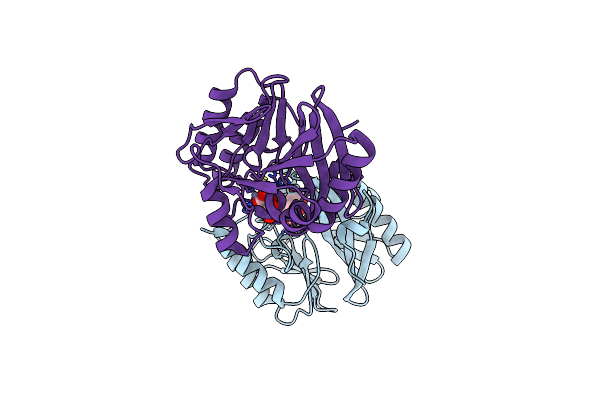 |
Crystal Structure Of The Di-Zinc Metallo-Beta-Lactamase Vim-4 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: ZN, FLC |
 |
Structural Basis For The Broad-Spectrum Inhibition Of Metallo-{Beta}-Lactamases: L1- Is38 Complex
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-28 Classification: HYDROLASE Ligands: ZN, SO4, I38 |
 |
Crystal Structure Of The Zinc Carbapenemase Cpha In Complex With The Inhibitor D-Captopril
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: ZN, SO4, MCO, GOL |

