Search Count: 13
 |
Crystal Structure Of Cd32B (Fc Gamma Receptor Iib) In Complex With Human Igg1 Fab Fragment (6G08)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-25 Classification: IMMUNE SYSTEM Ligands: NAG, GOL, PO4 |
 |
Organism: Bertholletia excelsa
Method: SOLUTION NMR Release Date: 2012-10-17 Classification: ALLERGEN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-06-10 Classification: HYDROLASE Ligands: CL |
 |
Deformation Of Helix C In The Low-Temperature L-Intermediate Of Bacteriorhodopsin
Organism: Halobacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-04-06 Classification: TRANSPORT PROTEIN Ligands: RET |
 |
Organism: Camelus dromedarius, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2003-10-14 Classification: HYDROLASE |
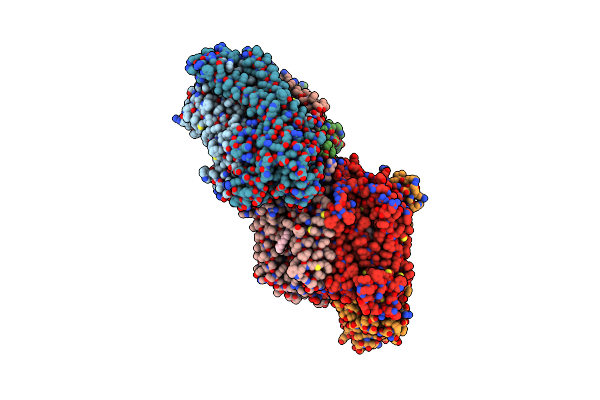 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-08-28 Classification: OXIDOREDUCTASE Ligands: CU, MG, CA, HEA, 3PE |
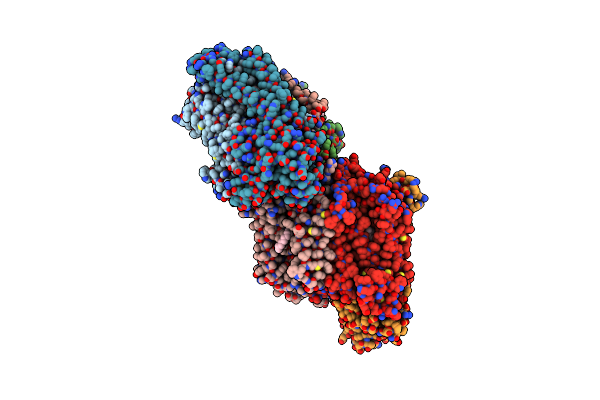 |
Structure Of Cytochrome C Oxidase From Rhodobacter Sphaeroides (Eq(I-286) Mutant))
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-08-28 Classification: OXIDOREDUCTASE Ligands: CU, MG, CA, HEA, 3PE |
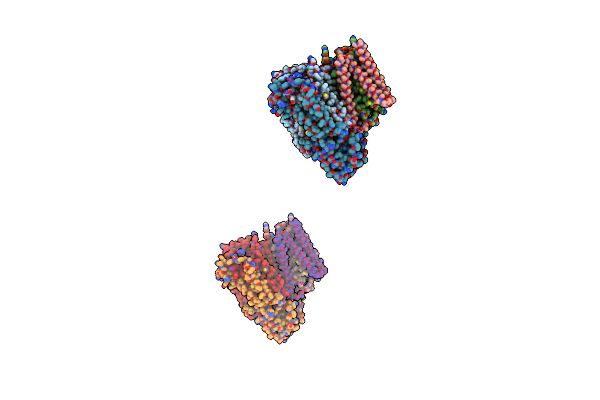 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2000-10-18 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HEO |
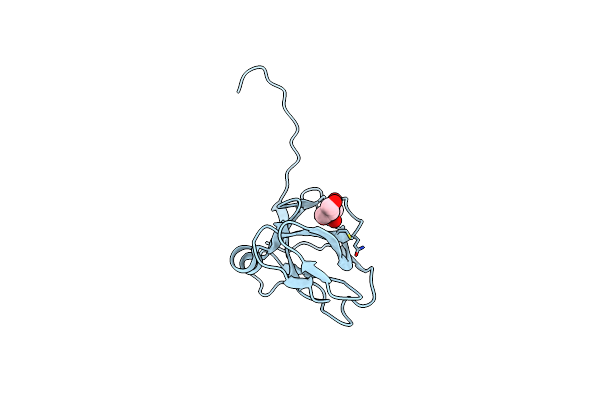 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2000-05-03 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2000-05-03 Classification: HYDROLASE Ligands: EMC, GOL |
 |
The Nmr Solution Structure Of The Homeodomain Of The Rat Insulin Gene Enhancer Protein Isl-1, 50 Structures
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 1999-06-15 Classification: DNA BINDING PROTEIN |
 |
Deoxyuridine 5'-Triphosphate Nucleotide Hydrolase (D-Utpase) Complexed With The Substrate Analogue Deoxyuridine 5'-Diphosphate (D-Udp)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1996-11-08 Classification: HYDROLASE Ligands: DUD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-11-14 Classification: HYDROLASE |

