Search Count: 6
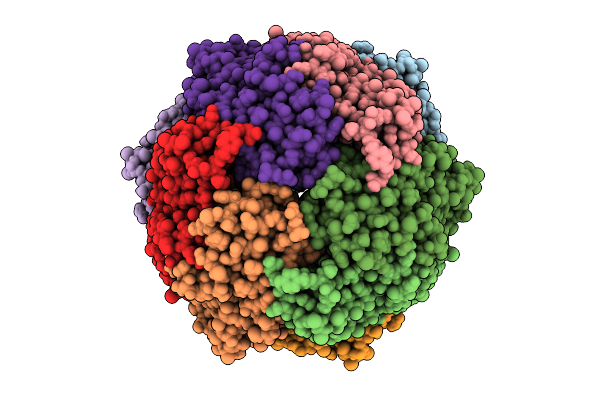 |
Structure Of E. Coli Dna Protection During Starvation Protein (Dps) From Single Particle Cryoem
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: FE |
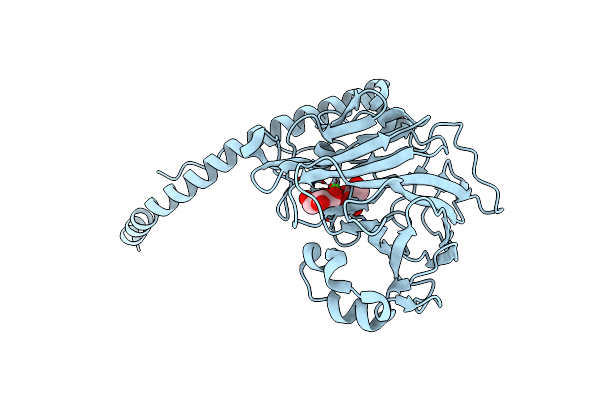 |
Glucan Binding Protein C Of Streptococcus Mutans Mediates Both Sucrose-Independent And Sucrose-Dependent Adherence
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-05-09 Classification: SUGAR BINDING PROTEIN Ligands: CA, BGC |
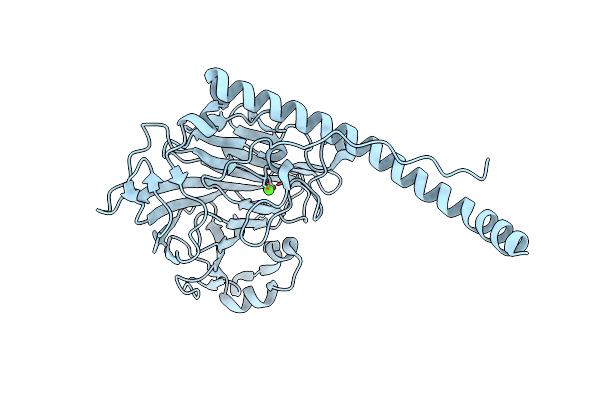 |
Structural Analysis Of The Glucan Binding Protein C Of Streptococcus Mutans Provides Evidence That It Mediates Both Sucrose-Independent And -Dependent Adherence
Organism: Streptococcus mutans ua159
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-03-07 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-04-20 Classification: CELL ADHESION Ligands: CA, MG, GLC |
 |
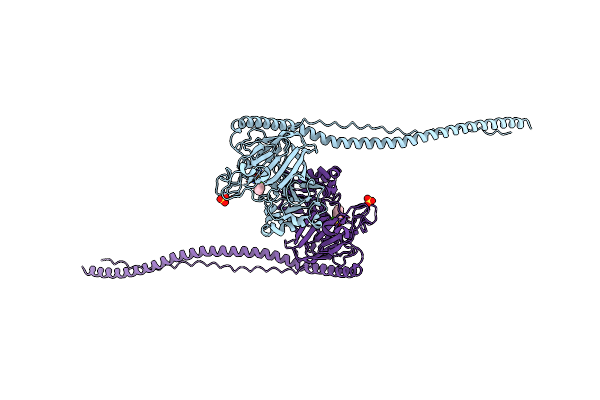
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-04-14 Classification: CELL ADHESION Ligands: PMS, SO3, CA |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2010-03-31 Classification: CELL ADHESION Ligands: PMS, SO4, CA |

