Search Count: 15
 |
Dna Replication Fork Binding Triggers Structural Changes In The Pria Dna Helicase That Regulate The Pria-Prib Replication Restart Pathway In E. Coli
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: HELICASE/DNA Ligands: ZN |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: FES, FE |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: FES, SO4, UB7, FE |
 |
Organism: Comamonas sp., Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: SO4, FES, FE, 8IB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-25 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-03-25 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Puta86-630 Mutant D370A Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, 1PE |
 |
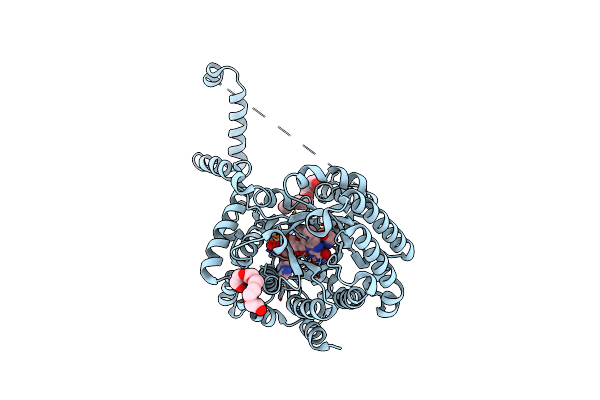
Crystal Structure Of Puta86-630 Mutant D370N Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, 1PE |
 |
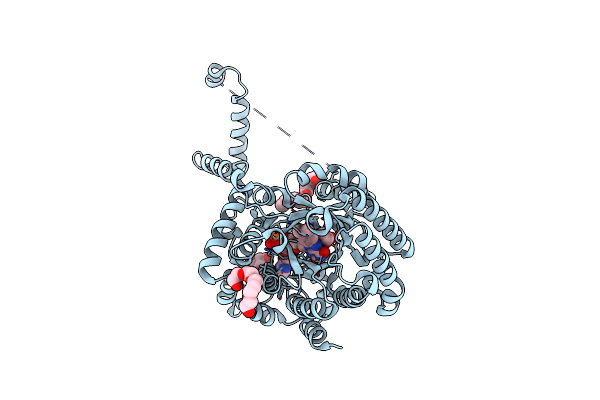
Crystal Structure Reduced Puta86-630 Mutant Y540S Complexed With Trans-4-Hydroxy-L-Proline
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, HYP, 1PE |
 |
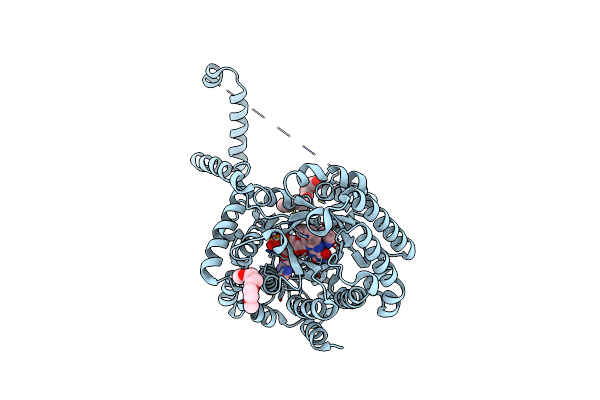
Crystal Structure Puta86-630 Mutant Y540S Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, CIT, 1PE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-02-03 Classification: OXIDOREDUCTASE Ligands: PRO, FAD, 1PE |
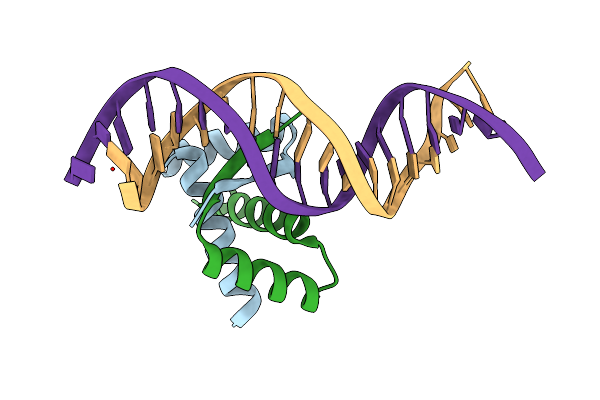 |
Structure Of The Ribbon-Helix-Helix Domain Of Escherichia Coli Puta (Puta52) Complexed With Operator Dna (O2)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2008-07-29 Classification: OXIDOREDUCTASE/DNA |
 |
Structure Of The Dna-Binding Domain Of E. Coli Proline Utilization A (Puta)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-02-20 Classification: DNA BINDING PROTEIN Ligands: IMD |
 |
Structure Of The Lys9Met Mutant Of The E. Coli Proline Utilization A (Puta) Dna-Binding Domain.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-08-15 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: CALCIUM-BINDING PROTEIN Ligands: CA, SO4 |

