Search Count: 7
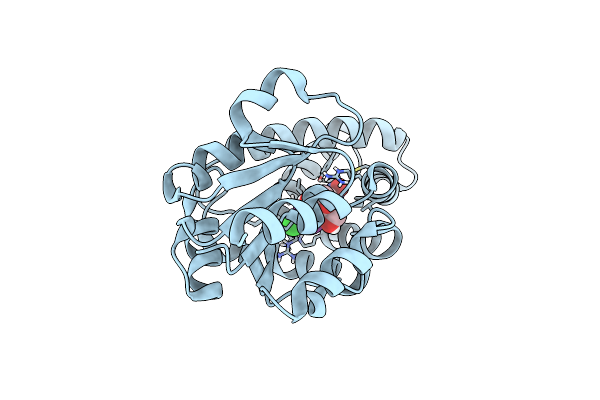 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, A1IGD, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD, PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, PEG, CL, PGE |
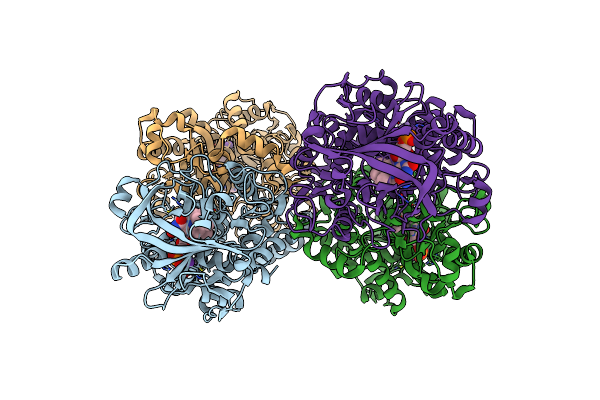 |
Structure Of The Reversible Pyrrole-2-Carboxylic Acid Decarboxylase Pa0254/Huda
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-04-07 Classification: LIGASE Ligands: MN, K, 4LU, IMD |
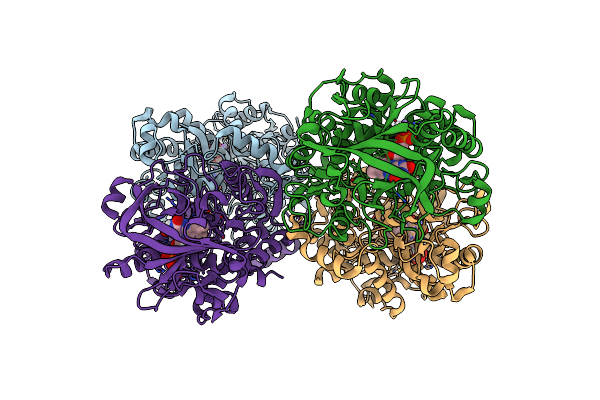 |
Structure Of The N318H Variant Of The Reversible Pyrrole-2-Carboxylic Acid Decarboxylase Pa0254/Huda In Complex With Fmn
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-04-07 Classification: LIGASE Ligands: FMN, MN, NA |
 |
Structure Of The Ubid-Class Enzyme Hmff From Pelotomaculum Thermopropionicum In Complex With Fmn
Organism: Pelotomaculum thermopropionicum si
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: FMN, K, MN, CA |
 |
Structure Of An Evolved Dimeric Form Of The Ubid-Class Enzyme Hmff From Pelotomaculum Thermopropionicum In Complex With Prfmn
Organism: Pelotomaculum thermopropionicum (strain dsm 13744 / jcm 10971 / si)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: K, MN, 4LU, CA |

