Search Count: 36
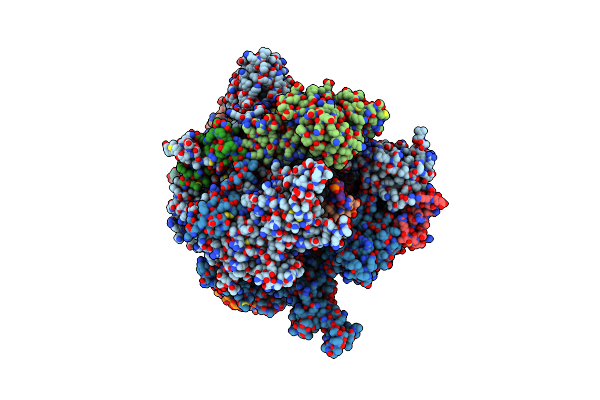 |
Architecture Of The Rna Polymerase Ii-Mediator Core Transcription Initiation Complex
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:6.60 Å Release Date: 2015-02-04 Classification: TRANSCRIPTION Ligands: ZN, MG |
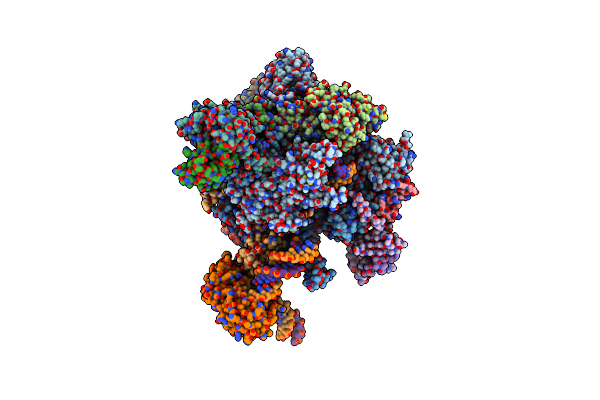 |
Architecture Of The Rna Polymerase Ii-Mediator Core Transcription Initiation Complex
Organism: Saccharomyces cerevisiae, Saccharomyces mikatae, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:7.80 Å Release Date: 2015-02-04 Classification: TRANSCRIPTION Ligands: ZN, MG |
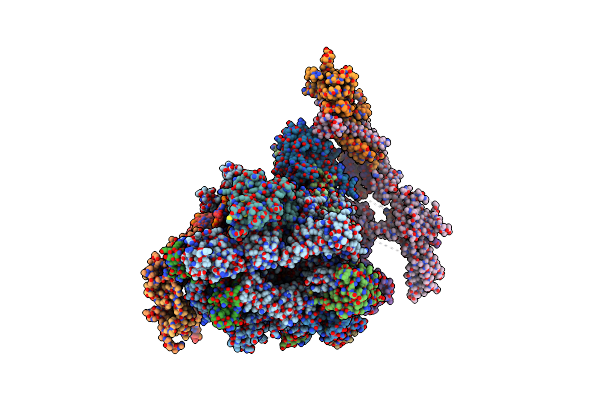 |
Architecture Of The Rna Polymerase Ii-Mediator Core Transcription Initiation Complex
Organism: Saccharomyces cerevisiae, Saccharomyces mikatae, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:9.70 Å Release Date: 2015-02-04 Classification: TRANSCRIPTION Ligands: ZN, MG |
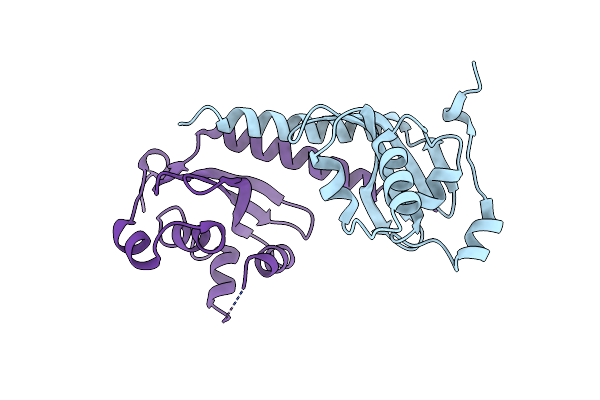 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-10-31 Classification: TRANSCRIPTION |
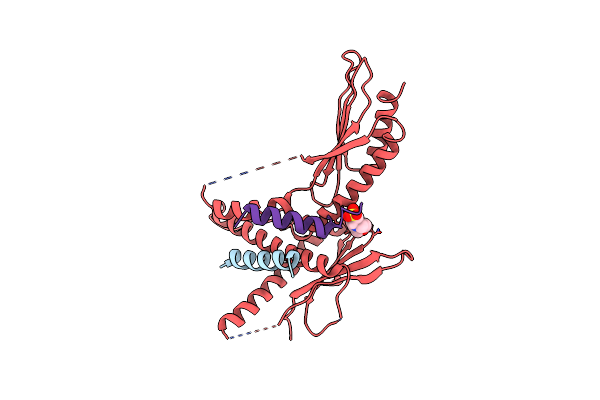 |
Structure Of The Saccharomyces Cerevisiae Mediator Subcomplex Med17C/Med11C/Med22C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-10-31 Classification: TRANSCRIPTION Ligands: MES |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2012-10-31 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-04-27 Classification: TRANSCRIPTION |
 |
The Mediator Med25 Activator Interaction Domain: Structure And Cooperative Binding Of Vp16 Subdomains
|
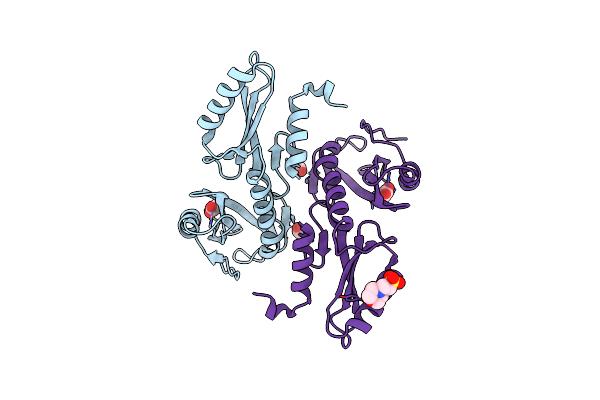 |
A Tandem Sh2 Domain In Transcription Elongation Factor Spt6 Binds The Phosphorylated Rna Polymerase Ii C-Terminal Repeat Domain(Ctd)
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-11-24 Classification: TRANSCRIPTION Ligands: ACT, MES |
 |
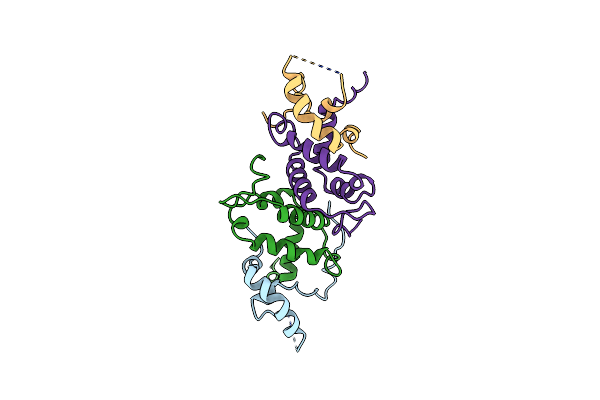
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-12-16 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2008-12-16 Classification: TRANSCRIPTION |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-04-22 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Yfh7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase With A Circular Permutation.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-03-27 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of Yfh7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase With A Circular Permutation.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2007-03-20 Classification: UNKNOWN FUNCTION Ligands: HG, SO4, BME |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-09-12 Classification: TRANSCRIPTION Ligands: PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-09-12 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-06-27 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-08-30 Classification: TRANSFERASE Ligands: CL, UDP, EDO, GOL |
 |
Alpha-Glucosyltransferase In Complex With Udp-Glucose And Dna Containing An Abasic Site
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-08-30 Classification: TRANSFERASE/DNA Ligands: UPG, GOL, PEG, UDP |
 |
Alpha-Glucosyltransferase In Complex With Udp And A 13_Mer Dna Containing A Hmu Base At 2.8 A Resolution
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-08-30 Classification: TRANSFERASE/DNA Ligands: NCO, UDP, CL, EDO |

