Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: IMMUNE SYSTEM Ligands: GOL, NA, CL, NAG |
 |
G169V Variant Of Streptomyces Coelicolor Coproheme Decarboxylase In Complex With Monovinyl, Monopropionate Deuteroheme
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: VOV, PO4, NA |
 |
G169L Variant Of Coproheme Decarboxylase From Streptomyces Coelicolor In Complex With Monovinyl, Monopropionate Deuteroheme
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: VOV |
 |
Wild-Type Coproheme Decarboxylase From Streptomyces Coelicolor In Complex With Monovinyl, Monopropionate Deuteroheme
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: VOV |
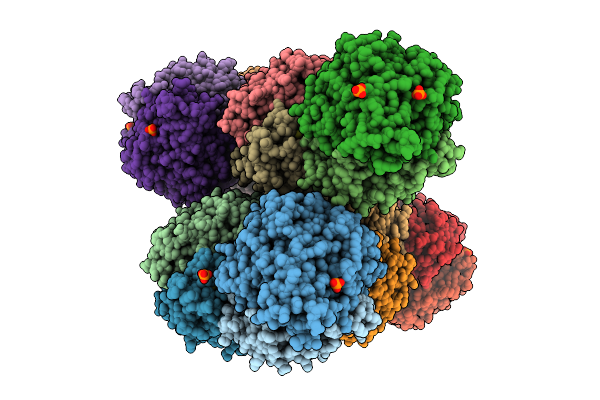 |
Alpha-Delta Heterodimeric Form Of Soluble Hydrogenase I From Pyrococcus Furiosus. Data Processed And Model Refined In P1
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE Ligands: SF4, FCO, NI, PO4, MG |
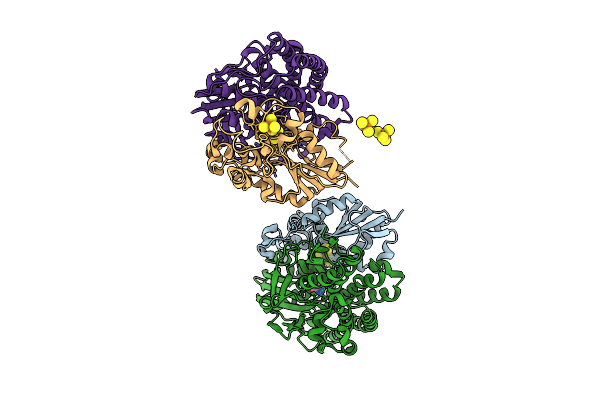 |
Alpha-Delta Heterodimeric Form Of Soluble Hydrogenase I From Pyrococcus Furiosus. Data Processed And Model Refined In P21221
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE Ligands: SF4, FCO, NI, CL |
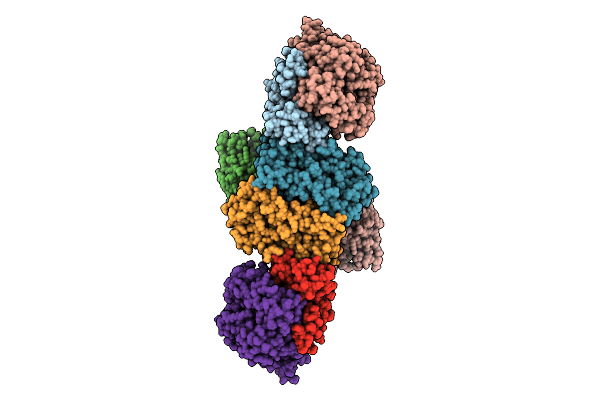 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FAD, NFU |
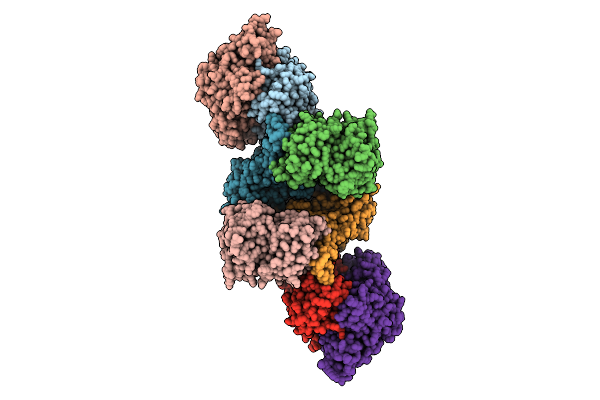 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FAD, NAP, NFU, MG |
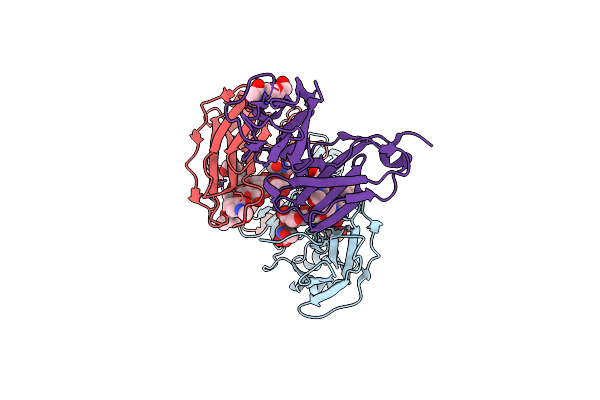 |
Crystal Structure Of The Low Affinity Fc Gamma Receptor Iiia Variant In Complex With The Fc Of Igg1.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-04-26 Classification: IMMUNE SYSTEM Ligands: NAG |
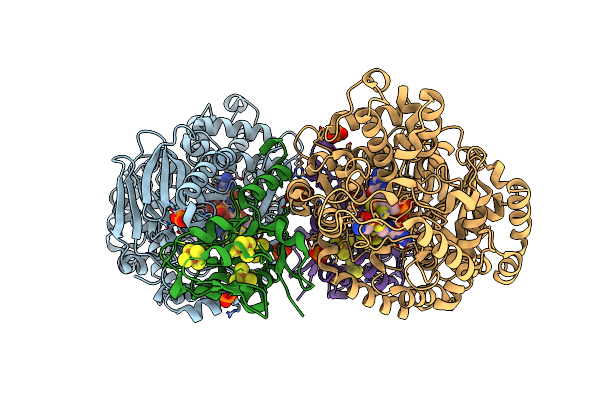 |
Organism: Pyrococcus furiosus com1
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-12-15 Classification: CYTOSOLIC PROTEIN Ligands: PTE, SF4, MG, TAU, PO4, CL |
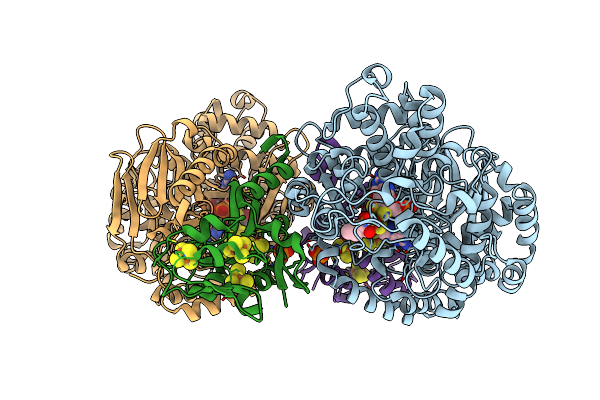 |
Organism: Pyrococcus furiosus com1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-11-24 Classification: OXIDOREDUCTASE Ligands: UKM, PTE, SF4, MG, PO4, CL |
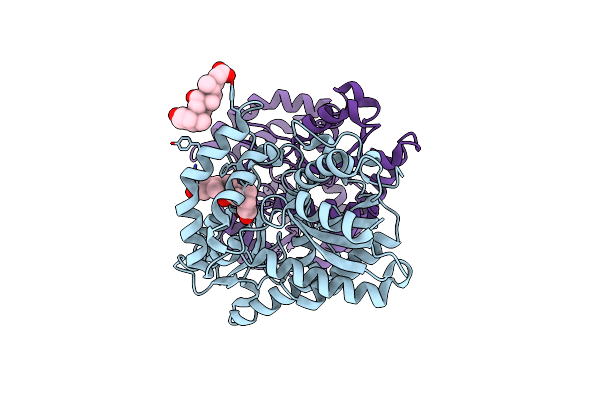 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-11-24 Classification: LYASE Ligands: CHD |
 |
Crystal Structures Of Beta-1,4-N-Acetylglucosaminyltransferase 2 (Pomgnt2): Structural Basis For Inherited Muscular Dystrophies
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: UDP, NAG, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: NAG, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-09-09 Classification: METAL BINDING PROTEIN Ligands: HEM |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-09 Classification: TRANSPORT PROTEIN Ligands: PO4, IMD, NA |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: UNKNOWN FUNCTION Ligands: PO4, BU1 |
 |
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-17 Classification: LYASE Ligands: FE |
 |
1,2-Propanediol Dehydration In Roseburia Inulinivorans; Structural Basis For Substrate And Enantiomer Selectivity
Organism: Roseburia inulinivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-01 Classification: LYASE Ligands: EDO, PEG |
 |
1,2-Propanediol Dehydration In Roseburia Inulinivorans; Structural Basis For Substrate And Enantiomer Selectivity
Organism: Roseburia inulinivorans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-06-01 Classification: LYASE Ligands: PGO |